Whole Transcriptome Sequencing – Illumina
Features
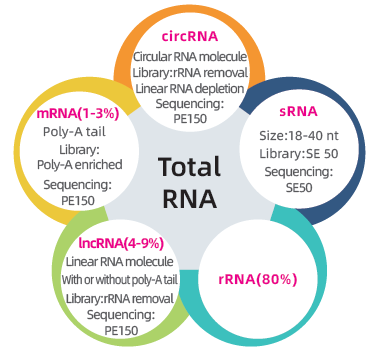
● Dual library to sequence the complete transcriptome: rRNA depletion followed by PE150 library preparation and size selection followed by SE50 library preparation
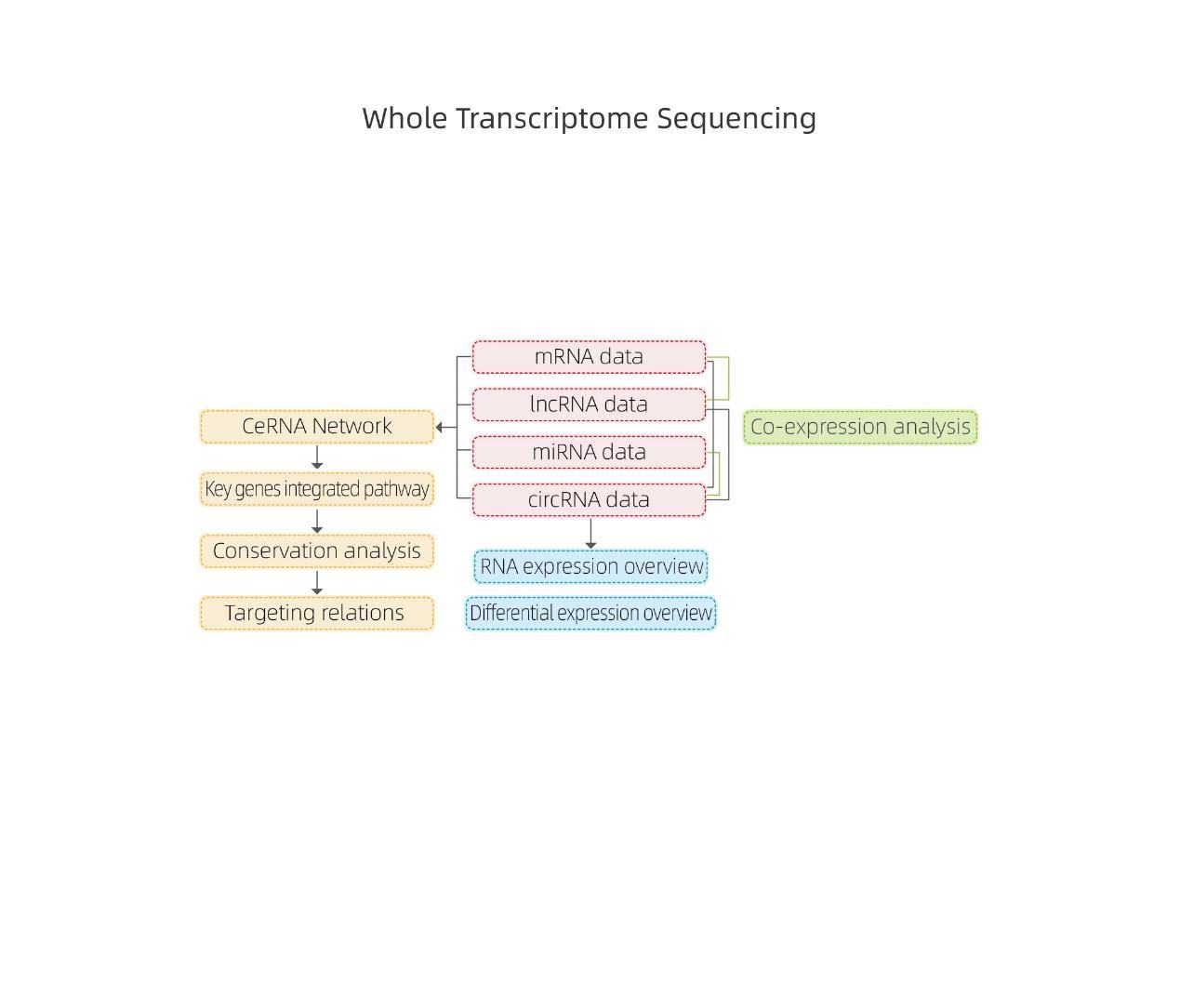
● Complete bioinformatics analysis of mRNA, lncRNA, circRNA, and miRNA in separate bioinformatics reports
● Joint analysis of all RNA expression in a combined report, including ceRNA networks analysis.
Service Advantages
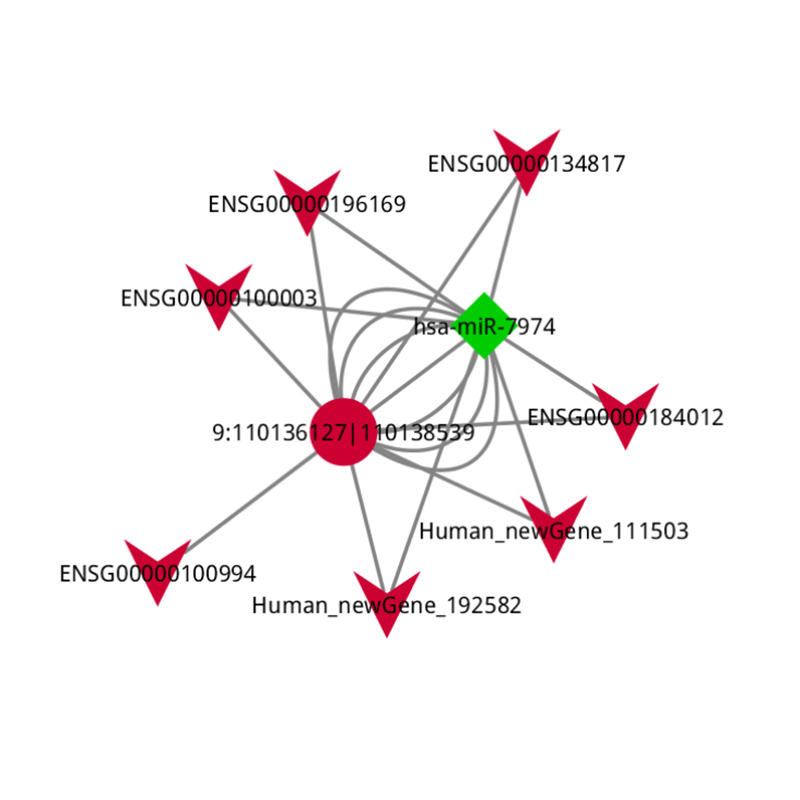
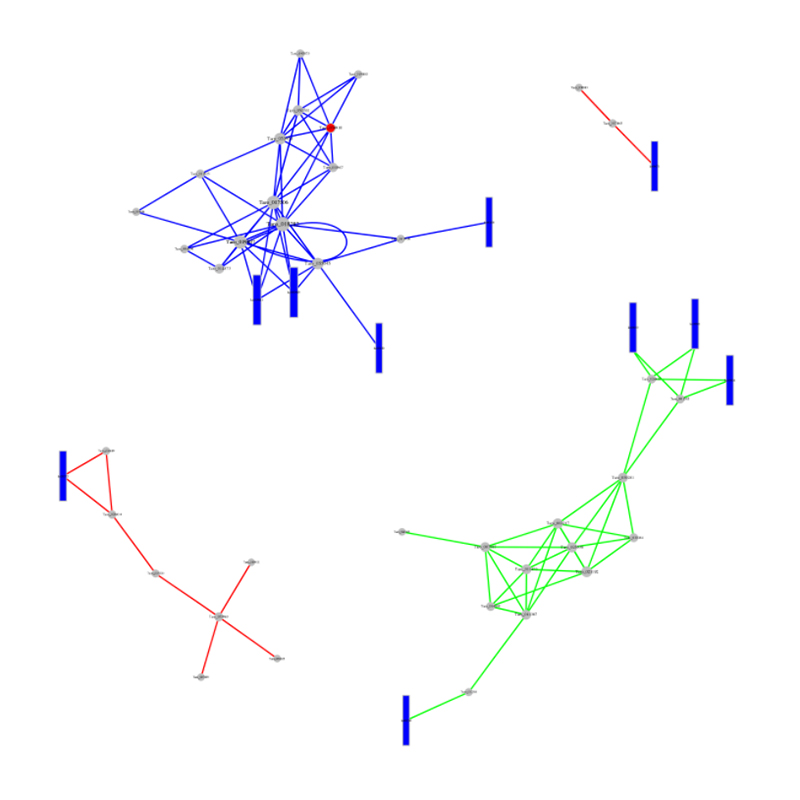
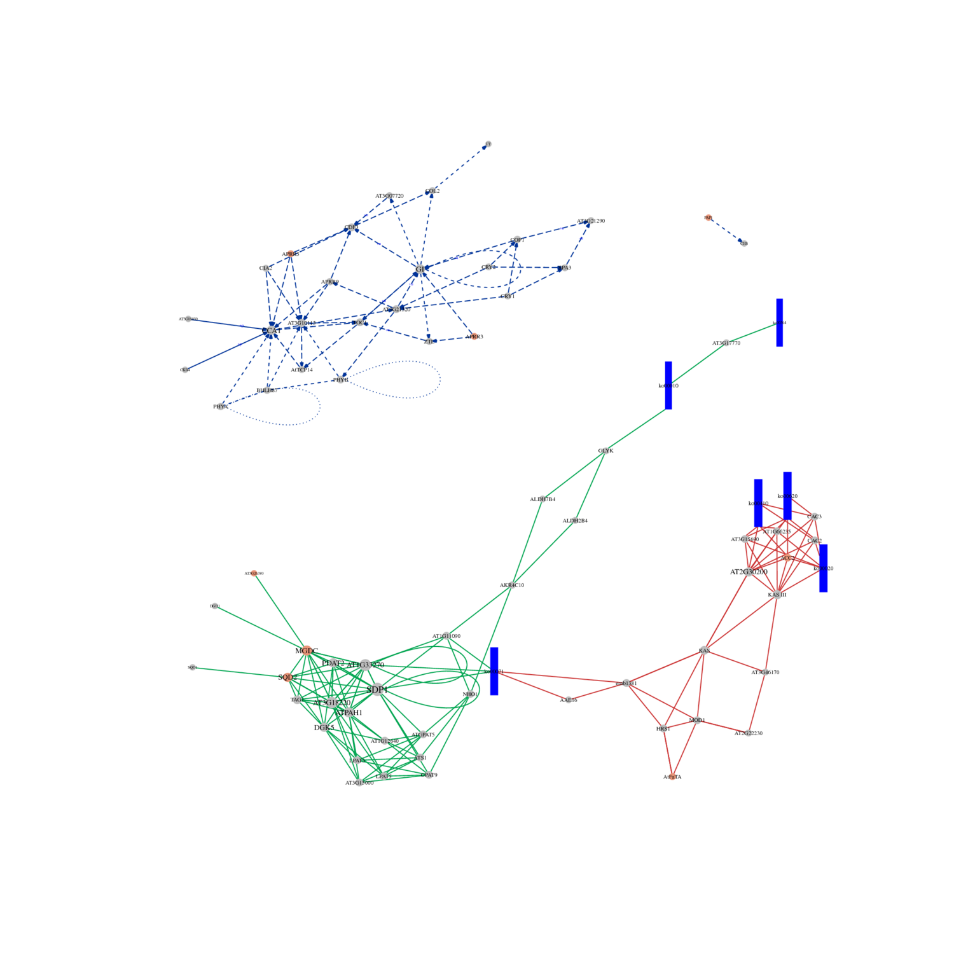
● In-depth Analysis of Regulatory Networks: ceRNA network analysis is enabled by the joint sequencing of mRNA, lncRNA, circRNA, and miRNA and by an exhaustive bioinformatic workflow.
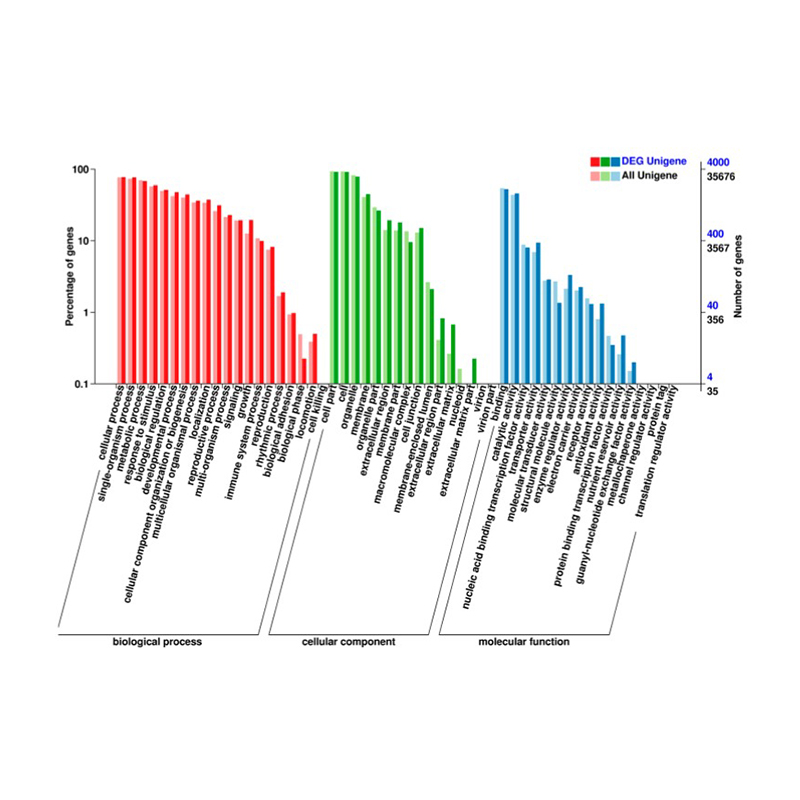
● Comprehensive Annotation: We use multiple databases to functionally annotate the Differentially Expressed Genes (DEGs) and perform the corresponding enrichment analysis, providing insights into the cellular and molecular processes underlying the transcriptome response.
● Extensive Expertise: With a track record of successfully closing over 2000 whole transcriptome projects in various research domain, our team brings a wealth of experience to every project.
● Rigorous Quality Control: We implement core control points across all stages, from sample and library preparation to sequencing and bioinformatics. This meticulous monitoring ensures the delivery of consistently high-quality results.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results
Sample Requirements and Delivery
|
Library |
Sequencing strategy |
Data recommended |
Quality Control |
|
rRNA depleted |
Illumina PE150 |
16 Gb |
Q30≥85% |
|
Size selected |
Illumina SE50 |
10-20M reads |
Sample Requirements:
Nucleotides:
|
Conc.(ng/μl) |
Amount (μg) |
Purity |
Integrity |
|
≥ 100 |
≥ 1 |
OD260/280=1.7-2.5 OD260/230=0.5-2.5 Limited or no protein or DNA contamination shown on gel. |
Plants: RIN≥6.5 Animal: RIN≥7.0 5.0≥28S/18S≥1.0; limited or no baseline elevation |
Recommended Sample Delivery
Container: 2 ml centrifuge tube (Tin foil is not recommended)
Sample labeling: Group+replicate e.g. A1, A2, A3; B1, B2, B3.
Shipment:
1. Dry-ice: Samples need to be packed in bags and buried in dry-ice.
2. RNAstable tubes: RNA samples can be dried in RNA stabilization tube(e.g. RNAstable®) and shipped at room temperature.
Service Work Flow

Experiment design

Sample delivery

RNA extraction

Library construction

Sequencing

Data analysis

After-sale services
Bioinformatics
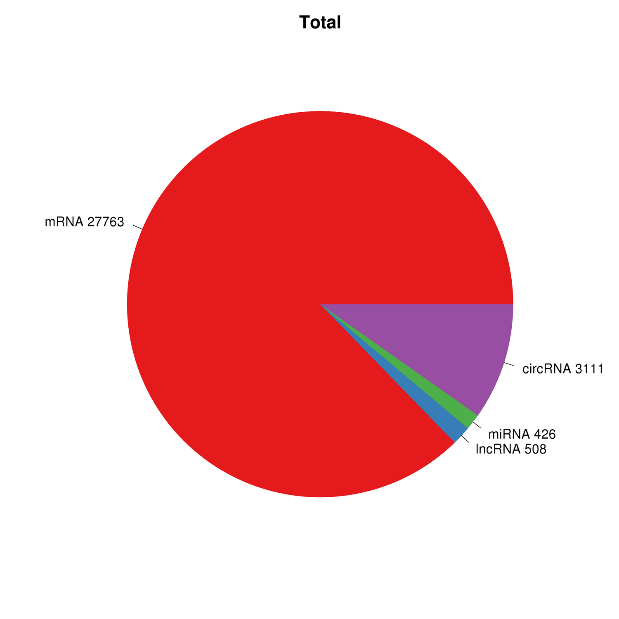
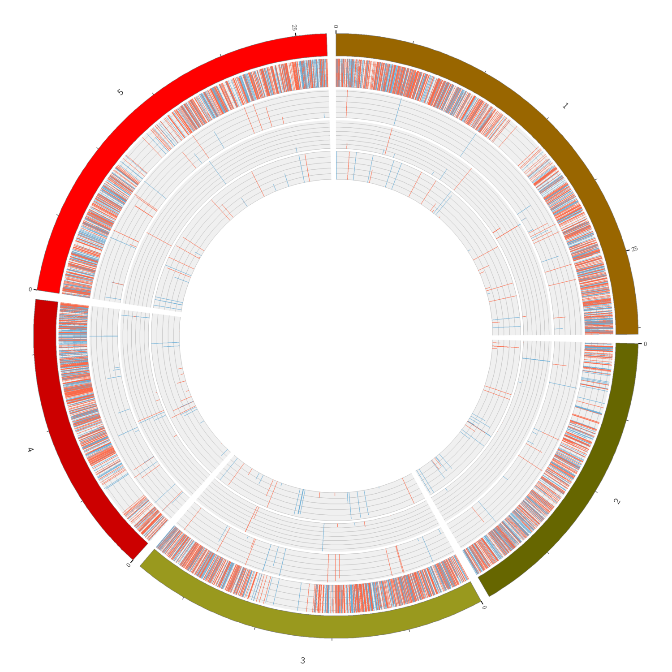
RNA expression overview
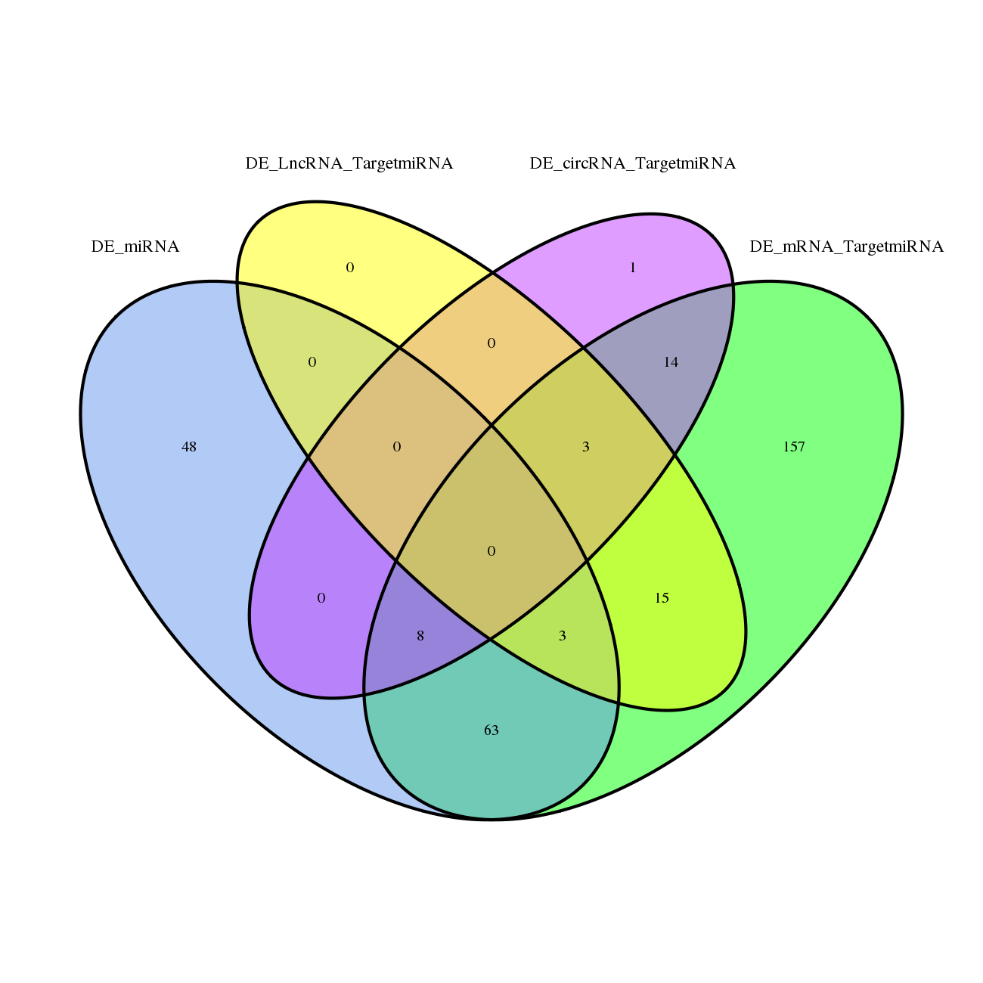
Differentially Expressed genes
ceRNA analysis
Explore the research advancements facilitated by BMKGene’ whole transcriptome sequencing services through a curated collection of publications.
Dai, Y. et al. (2022) ‘Comprehensive expression profiles of mRNAs, lncRNAs and miRNAs in Kashin-Beck disease identified by RNA-sequencing’, Molecular Omics, 18(2), pp. 154–166. doi: 10.1039/D1MO00370D.
Liu, N. nan et al. (2022) ‘Full length transcriptomes analysis of cold-resistance of Apis cerana in Changbai Mountain during overwintering period.’, Gene, 830, pp. 146503–146503. doi: 10.1016/J.GENE.2022.146503.
Wang, X. J. et al. (2022) ‘Multi-Omics Integration-Based Prioritisation of Competing Endogenous RNA Regulation Networks in Small Cell Lung Cancer: Molecular Characteristics and Drug Candidates’, Frontiers in Oncology, 12, p. 904865. doi: 10.3389/FONC.2022.904865/BIBTEX.
Xu, P. et al. (2022) ‘Integrated analysis of the lncRNA/circRNA-miRNA-mRNA expression profiles reveals novel insights into potential mechanisms in response to root-knot nematodes in peanut’, BMC Genomics, 23(1), pp. 1–12. doi: 10.1186/S12864-022-08470-3/FIGURES/7.
Yan, Z. et al. (2022) ‘Whole-transcriptome RNA sequencing highlights the molecular mechanisms associated with the maintenance of postharvest quality in broccoli by red LED irradiation’, Postharvest Biology and Technology, 188, p. 111878. doi: 10.1016/J.POSTHARVBIO.2022.111878.