T2T génom majelis, GAP FREE Génom
1stDua Génom Béas1
Judul: Majelis sareng Validasi Dua Génom Rujukan Gap-gratis pikeun Padi Xian/indica Ngungkabkeun Wawasan ngeunaan Arsitéktur Centromere Tutuwuhan
Doi:https://doi.org/10.1101/2020.12.24.424073
Dipasang waktos: 01 Januari 2021.
Institute: Universitas Pertanian Huazhong, Cina
Bahan
O. sativa xian / indicavariétas béas 'Zhenshan 97 (ZS97)' jeung 'Minghui 63 (MH63)
Stratégi sequencing
NGS dibaca + HiFi dibaca + CLR dibaca + BioNano + Hi-C
Data:
ZS97: 8,34 Gb(~23x)HiFi dibaca + 48,39 Gb (~131x) CLR dibaca + 25 Gb(~69x) NGS + 2 sél BioNano Irys
MH63: 37,88 Gb (~103x) HiFi dibaca + 48,97 Gb (~132x) CLR dibaca + 28 Gb(~76x) NGS + 2 sél BioNano Irys
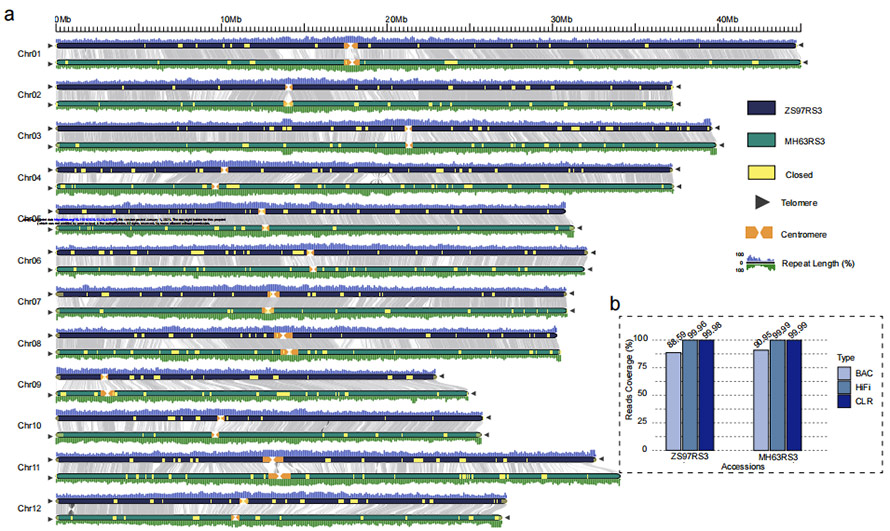
Gambar 1 Dua génom béas bébas gap (MH63 jeung ZS97)
2ndGénom pisang2
Judul: Telomere-to-telomere kromosom gapless pisang ngagunakeun urutan nanopore
Doi:https://doi.org/10.1101/2021.04.16.440017
Dipasang waktos: April 17, 2021.
Institut: Université Paris-Saclay, Perancis
Bahan
Ganda haploidMusa acuminatasppmalaccensis(DH-Pahang)
Strategi sequencing sareng data:
Modeu HiSeq2500 PE250 + Minion/PromethION (93Gb,~200X)+ Peta optik (DLE-1+BspQ1)
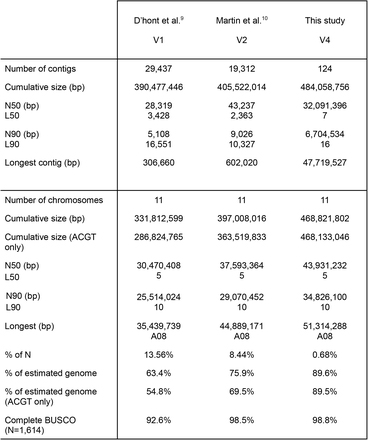
meja 1 Babandingan Musa acuminata (DH-Pahang) rakitan génom
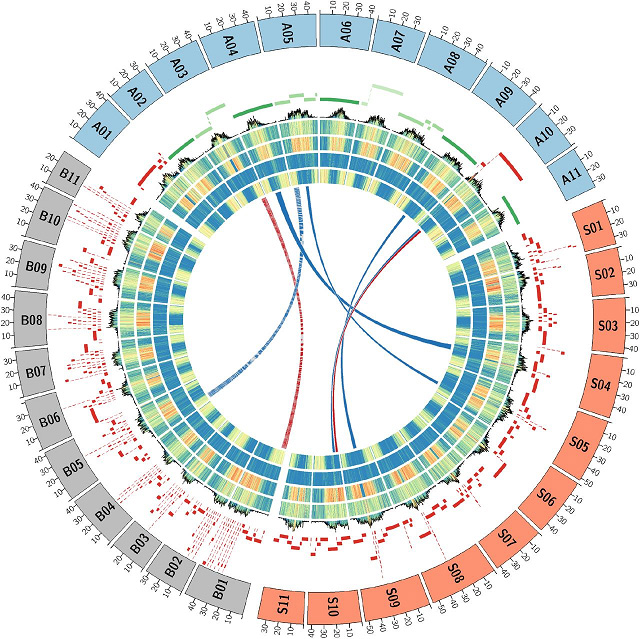
Gambar 2 Musa génoms arsitéktur ngabandingkeun
3rdgénom Phaeodactylum tricornutum3
Judul: Telomere-to-telomere génom assembly ofP
haeodactylum tricornutum
Doi:https://doi.org/10.1101/2021.05.04.442596
Dipasang waktos: 04 Mei 2021
Institute: Universitas Kulon, Kanada
Bahan
Phaeodactylum tricornutum(Koleksi Budaya Ganggang sareng Protozoa CCAP 1055/1)
Strategi sequencing sareng data:
1 sél aliran minION Oxford Nanopore + 2 × 75 dipasangkeun-tungtung pertengahan kaluaran NextSeq 550 ngajalankeun
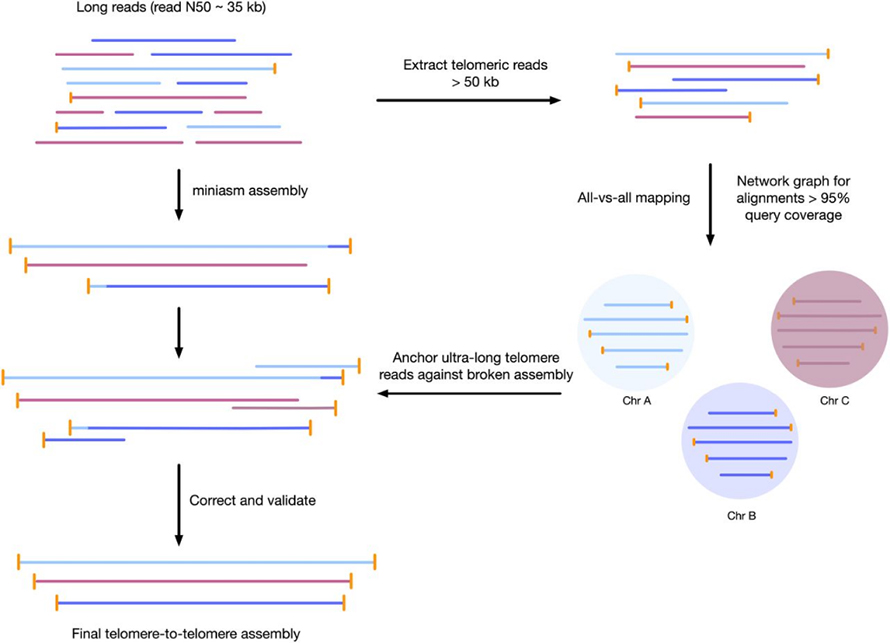
Gambar 3 Workflow pikeun rakitan génom telomere-to-telomere
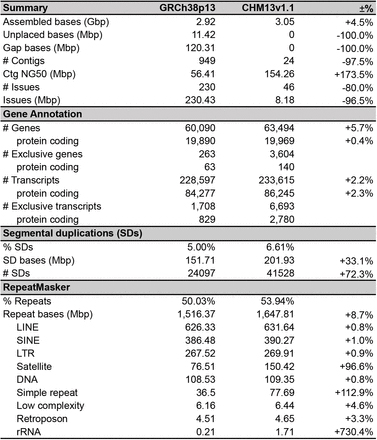
4thGénom CHM13 manusa4
Judul: Runtuyan lengkep génom manusa
Doi:https://doi.org/10.1101/2021.05.26.445798
Dipasang waktos: 27 Mei 2021
Institute: National Institutes of Kaséhatan (NIH), AS
Bahan: garis sél CHM13
Strategi sequencing sareng data:
30× Sequencing konsensus sirkular PacBio (HiFi), 120× Oxford Nanopore ultra-long read sequencing, 100× Illumina PCR-Free sequencing (ILMN), 70× Illumina / Arima Genomics Hi-C (Hi-C), peta optik BioNano, jeung Strand-seq
meja 2 Babandingan GRCh38 na T2T-CHM13 majelis génom manusa
Rujukan
1. Sergey Nurk et al.Runtuyan lengkep génom manusa.bioRxiv 2021.05.26.445798;doi:https://doi.org/10.1101/2021.05.26.445798
2.Caroline Belser dkk.Telomere-to-telomere kromosom gapless cau maké sequencing nanopore.bioRxiv 2021.04.16.440017;doi:https://doi.org/10.1101/2021.04.16.440017
3.Daniel J. Giguere dkk.Rakitan génom telomere-to-telomere tina Phaeodactylum tricornutum.bioRxiv 2021.05.04.442596;doi:https://doi.org/10.1101/2021.05.04.442596
4.Jia-Ming Song et al.Majelis sareng Validasi Dua Génom Rujukan Gap-gratis pikeun Xian / Indica Rice nembongkeun Wawasan kana Arsitéktur Centromere Plant.bioRxiv 2020.12.24.424073;doi:https://doi.org/10.1101/2020.12.24.424073
waktos pos: Jan-06-2022

