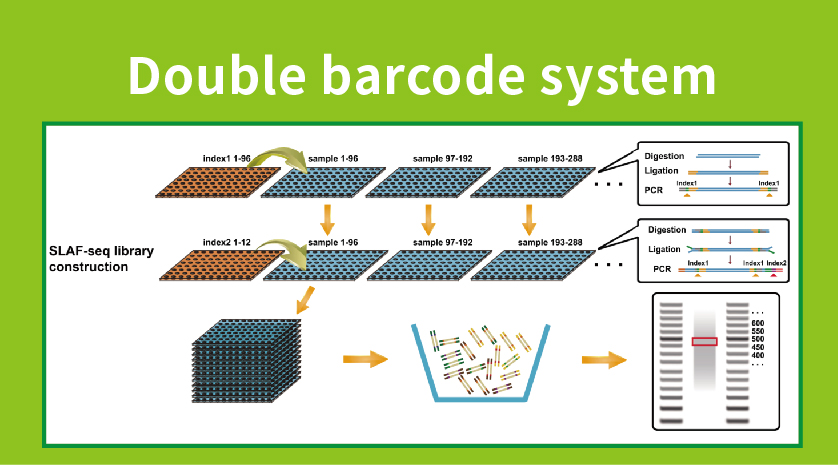
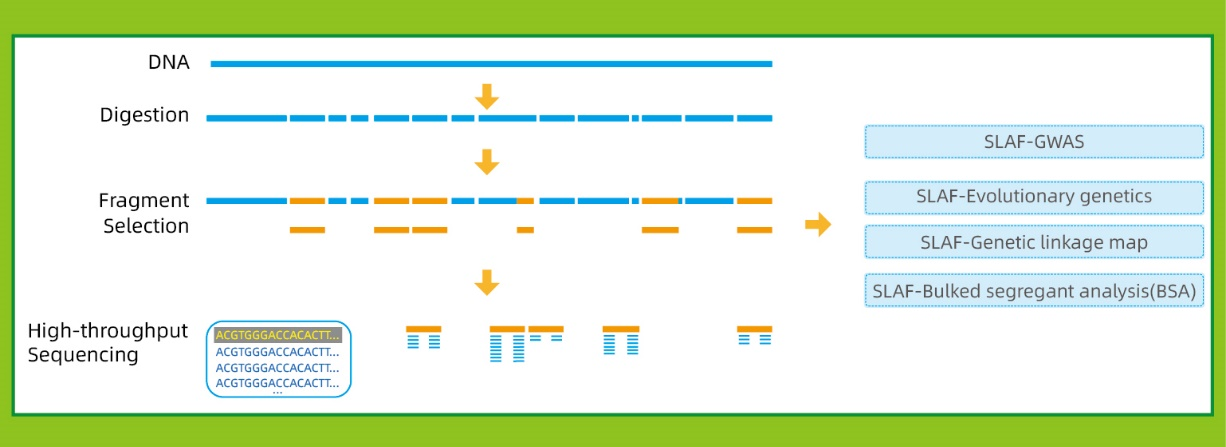
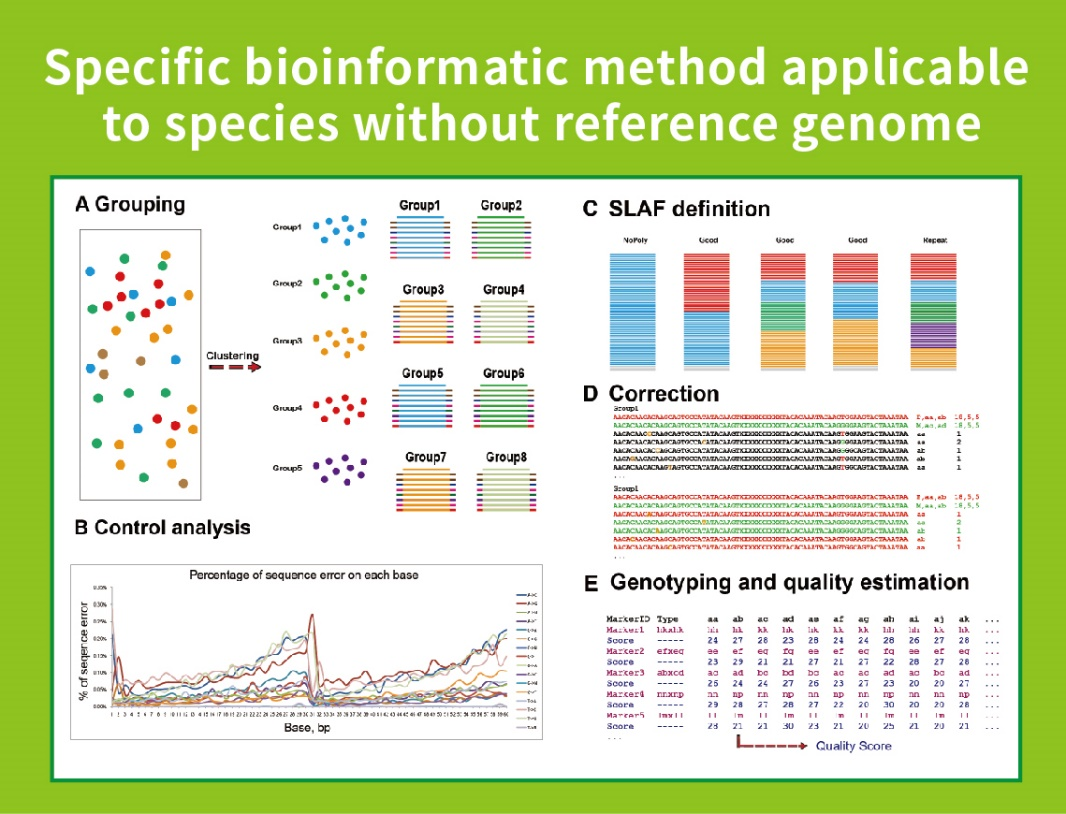
Specific-Locus Amplified Fragment Sequencing (SLAF-Seq)
Service Details
Technical Scheme
Kuyerera kwebasa

Basa Rinobatsira
Mucherechedzo wepamusoro wawanikwa-Yakakwira-yekuteedzera tekinoroji inobatsira SLAF-Seq mukuwana mazana ezviuru zvemategi mukati meiyo genome yese.
Kuderera kutsamira pane genome- Inogona kuiswa kune zvipenyu zvine kana isina referensi genome.
Flexible chirongwa dhizaini-Imwe enzyme, mbiri-enzyme, yakawanda-enzyme digestion uye akasiyana marudzi ee enzymes, ese anogona kusarudzwa kuti aite akasiyana tsvakiridzo chinangwa kana marudzi.Pre-evaluation musilico inoshandiswa kuvimbisa yakakwana enzyme dhizaini.
Kubudirira enzymatic digestion- Pre-kuyedza yakaitwa kuti ikwidzise mamiriro ezvinhu, izvo zvinoita kuti kuyedza kwakarongeka kugadzikane uye kwakavimbika.Fragment kuunganidza kushanda zvakanaka kunogona kuwana pamusoro pe95%.
Akagovaniswa ma tag eSLAF zvakaenzana-Ma tag eSLAF akagovaniswa zvakaenzana mumakromosomes ese kusvika pamwero mukuru, achiwana avhareji ye1 SLAF pa4 kb.
Kubudirira kudzivisa kudzokorora-Kudzokorora kutevedzana muSLAF-Seq data yakaderedzwa kusvika yakaderera kupfuura 5%, kunyanya mumhando dzine huwandu hwekudzokorora, senge gorosi, chibage, nezvimwe.
Zvakawanda ruzivo-Kupfuura 2000 yakavharwa mapurojekiti eSLAF-Seq pamazana emarudzi anofukidza zvidyarwa, mhuka dzinoyamwisa, shiri, zvipembenene, zvipenyu zvemumvura, nezvimwe.
Kuzvigadzira pachako bioinformamatic workflow-Iyo yakasanganiswa bioinformatic workflow yeSLAF-Seq yakagadziriswa neBMKGENE kuve nechokwadi chekuvimbika uye chokwadi chekubuda kwekupedzisira.
Sevhisi Zvinotsanangurwa
| Platform | Conc.(ng/gl) | Zvose (ug) | OD260/280 |
| Illumina NovaSeq | >35 | >1.6(Vhoriyamu> 15μl) | 1.6-2.5 |
Yakakurudzirwa Sequencing Strategy
Sequencing kudzika: 10X/Tag
| Kukura kweGenome | Yakakurudzirwa SLAF Tags |
| <500Mb | 100K kana WGS |
| 500 Mb- 1 Gb | 100 K |
| 1 Gb -2 Gb | 200 K |
| Giant kana yakaoma genomes | 300 - 400K |
| Applications
| Recommended Population Scale
| Sequencing strategy nekudzika
| |
| Kudzika
| Nhamba yeTag
| ||
| GWAS
| Nhamba yemuenzaniso ≥200
| 10X
|
Maererano ne genome size
|
| Genetic Evolution
| Munhu mumwe nomumwe boka duku ≥ 10; sampuli dzese ≥ 30
| 10X
| |
Yakakurudzirwa Sample Delivery
mudziyo: 2 ml centrifuge chubhu
Kune akawanda emasampuli, isu tinokurudzira kuti usachengetedze mu ethanol.
Sample labeling: Samples dzinofanirwa kunyorwa zvakajeka uye dzakafanana neyakatumirwa ruzivo ruzivo fomu.
Shipment: Dry-ice: Samples dzinofanirwa kurongedzerwa mumabhegi kutanga uye kuvigwa mune yakaoma-aizi.
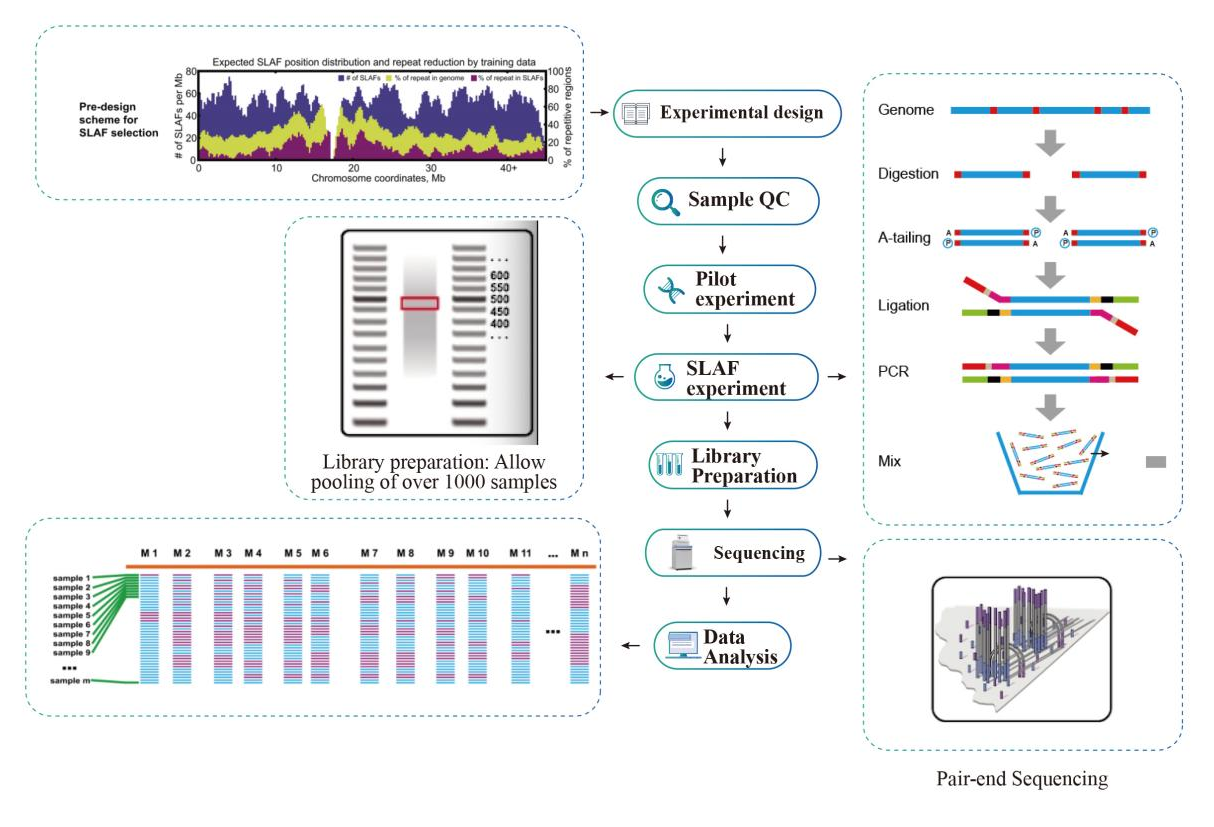
Service Workflow






Muenzaniso QC
Pilot experiment
SLAF-kuedza
Kugadzirira Raibhurari
Sequencing
Data Analysis
Mushure mekutengesa Services
1. Nhamba dzemhedzisiro yemepu
2. SLAF marker development
3. Kusiyana kwechirevo
| Gore | Journal | IF | Title | Applications |
| 2022 | Kutaurirana kwakasikwa | 17.694 | Genomic nheyo yegiga-chromosomes uye giga-genome yemuti peony Paeonia ostii | SLAF-GWAS |
| 2015 | New Phytologist | 7.433 | Domestication footprints anchor genomic regions of agronomic value in soya beans | SLAF-GWAS |
| 2022 | Chinyorwa che Advanced Research | 12.822 | Genome-wide artificial introgressions yeGossypium barbadense muG. hirsutum ratidza yepamusoro loci yekuvandudza panguva imwe chete yekotoni fiber mhando uye goho maitiro | SLAF-Evolutionary genetics |
| 2019 | Molecular Plant | 10.81 | Population Genomic Analysis uye De Novo Assembly Inozivisa Mavambo eWeedy Mupunga semutambo weEvolutionary | SLAF-Evolutionary genetics |
| 2019 | Nature Genetics | 31.616 | Genome sequence uye genetic diversity of the common carp, Cyprinus carpio | Mepu yeSLAF-Linkage |
| 2014 | Nature Genetics | 25.455 | Iyo genome yenzungu yakarimwa inopa nzwisiso mune legume karyotypes, polyploid. evolution uye kurima zvirimwa. | Mepu yeSLAF-Linkage |
| 2022 | Plant Biotechnology Journal | 9.803 | Kuzivikanwa kwe ST1 kunoratidza sarudzo inosanganisira kurovera kwembeu morphology uye zviri mukati memafuta panguva yekudyara soya bean | SLAF-Marker kuvandudza |
| 2022 | International Journal yeMolecular Sayenzi | 6.208 | Kuzivikanwa uye DNA Mucherechedzo Kukudziridzwa kweGorosi-Leymus mollis 2Ns (2D) Disomic Chromosome Kutsiva | SLAF-Marker kuvandudza |