
Chirimwa/Mhuka De Novo Genome Sequencing
Basa Rinobatsira
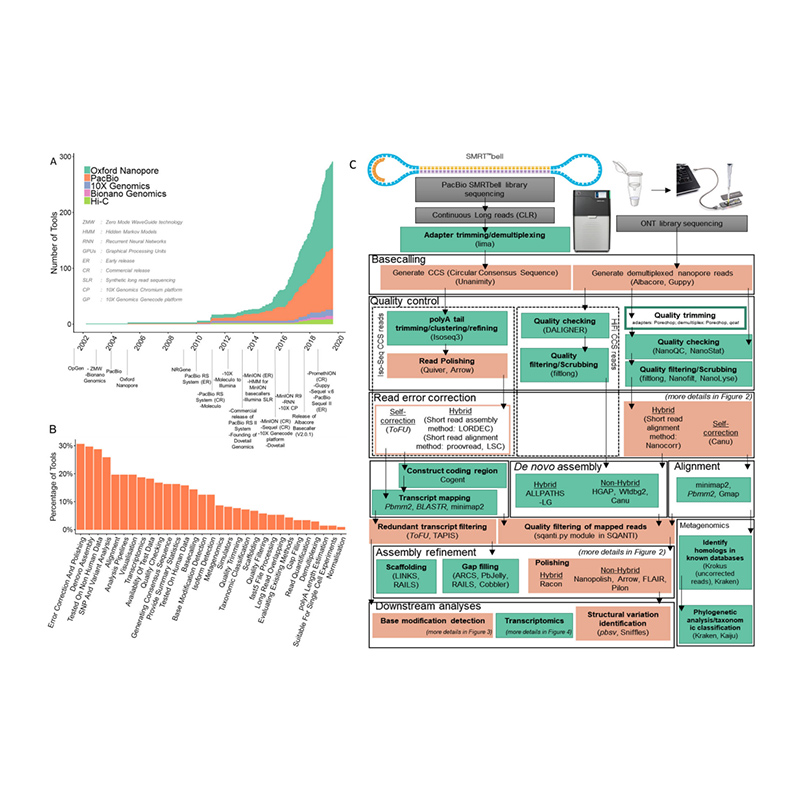
Kuvandudzwa kwemapuratifomu ekutevedzana uye bioinformatics mukatide novogenome assembly
(Amarasinghe SL et al.,Genome Biology, 2020)
● Kugadzira novel genomes uye kuvandudza mareferensi majenomes aripo emarudzi anofarira.
● Kururama kwepamusoro, kuenderera mberi uye kukwana mukuungana
● Kugadzira zviwanikwa zvakakosha zvekutsvaga mukutevedzana kwepolymorphism, QTLs, gene editing, kubereka, nezvimwe.
● Yakashongedzerwa nehuwandu hwakazara hwechitatu-chizvarwa chekutevedzana mapuratifomu: one-stop genome assembly solution.
● Kutevedzana kunochinjika uye kuunganidza mazano anozadzikisa majenome akasiyana ane maitiro akasiyana
● Chikwata chebioinformatician chine hunyanzvi chine ruzivo rwakakura mumagungano akaomarara ejenome, kusanganisira mapolyploid, magiant genome, nezvimwe.
● Mhosva dzinopfuura zana dzakabudirira dzine accumulative published impact factor inopfuura 900
● Kuchinja-chinja-nguva nekukurumidza semwedzi mitatu ye chromosome-level genome assembly.
● Rutsigiro rwehunyanzvi rwakasimba rune nhevedzano yematendi uye software yekodzero yekodzero mumativi ekuedza uye bioinformatics.
Sevhisi Zvinotsanangurwa
|
Content
|
Platform
|
Read Length
|
Coverage
|
| Genome Survey
| Illumina NovaSeq
| PE150
| ≥ 50X
|
| Genome Sequencing
| PacBio Revio
| 15 KB Inoverenga HiFi
| ≥ 30X
|
| Hi-C
| Illumina NovaSeq
| PE150
| ≥100X
|
Kuyerera kwebasa

Sample Zvinodiwa uye Delivery
Sample Zvinodiwa:
| Zvisikwa | Tissue | ZvePacBio | Pamusoro peNanopore |
| Mhuka | Visceral nhengo (chiropa, spleen, nezvimwewo) | ≥ 1.0 g | ≥ 3.5 g |
| Tsandanyama | ≥ 1.5 g | ≥ 5.0 g | |
| Ropa remhuka | ≥ 1.5 mL | ≥ 5.0 mL | |
| Ropa rehove kana shiri | ≥ 0.2 mL | ≥ 0.5 mL | |
| Zvirimwa | Mashizha matsva | ≥ 1.5 g | ≥ 5.0 g |
| Petal kana dzinde | ≥ 3.5 g | ≥ 10.0 g | |
| Midzi kana mhodzi | ≥ 7.0 g | ≥ 20.0 g | |
| Masero | Sero tsika | ≥ 3×107 | ≥ 1×108 |
Yakakurudzirwa Sample Delivery
Container: 2 ml centrifuge chubhu (Tin foil haina kukurudzirwa)
Kune akawanda emasampuli, isu tinokurudzira kuti usachengetedze mu ethanol.
Sample labeling: Samples dzinofanirwa kunyorwa zvakajeka uye dzakafanana neyakatumirwa ruzivo ruzivo fomu.
Shipment: Dry-ice: Samples dzinofanirwa kurongedzerwa mumabhegi kutanga uye kuvigwa mune yakaoma-aizi.
Kuyerera Kwebasa Rebasa

Kuedza kugadzira

Muenzaniso wekutumira

Kutorwa kweDNA

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
* Demo mhedzisiro inoratidzwa pano yese kubva kugenomes yakaburitswa neBiomarker Technologies
1.Circos pachromosome-level genome assembly yeG. rotundifoliumneNanopore sequencing chikuva
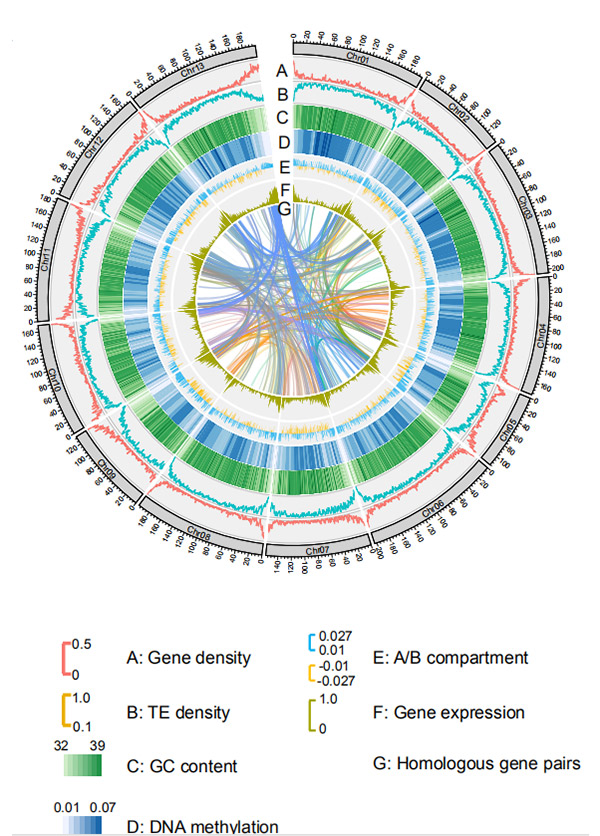
Wang M et al.,Molecular Biology uye Evolution, 2021
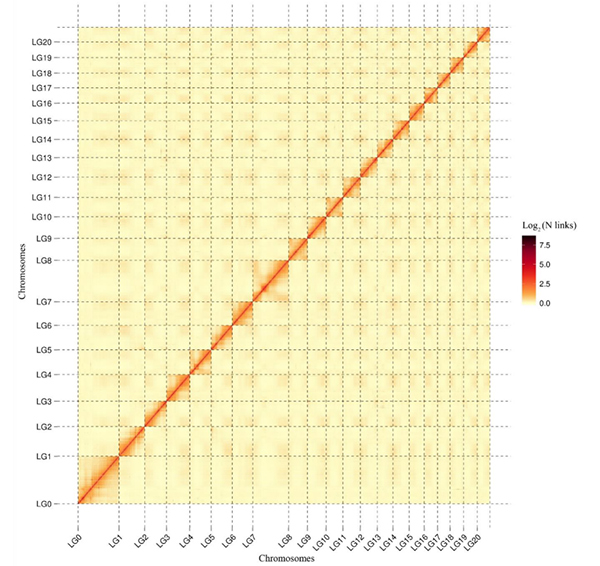
2.Statistics yeWeining rye genome assembly uye annotation
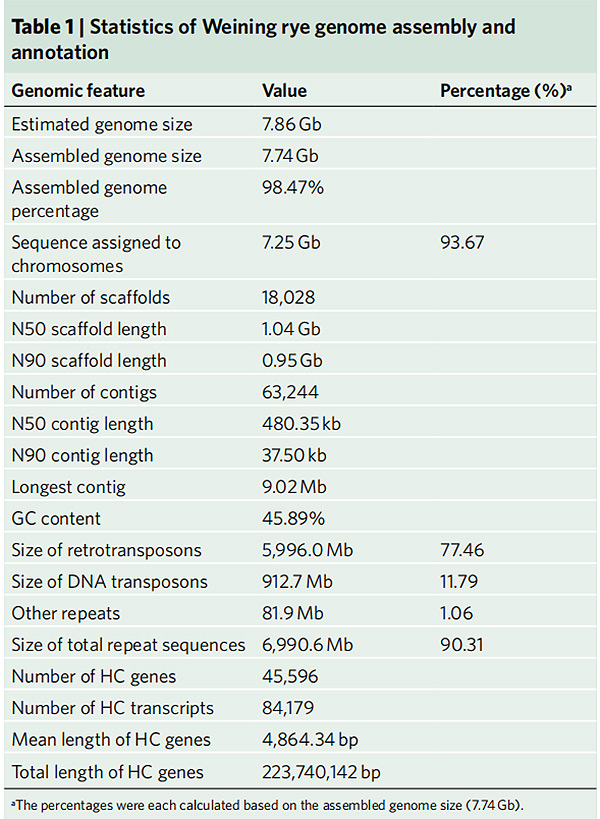
Li G et al.,Nature Genetics, 2021
3.Gene kufanotaura kweSechium edugenome, inotorwa kubva munzira nhatu dzekufanotaura:De novokufanotaura, Homology-yakavakirwa kufanotaura uye RNA-Seq data yakavakirwa kufanotaura
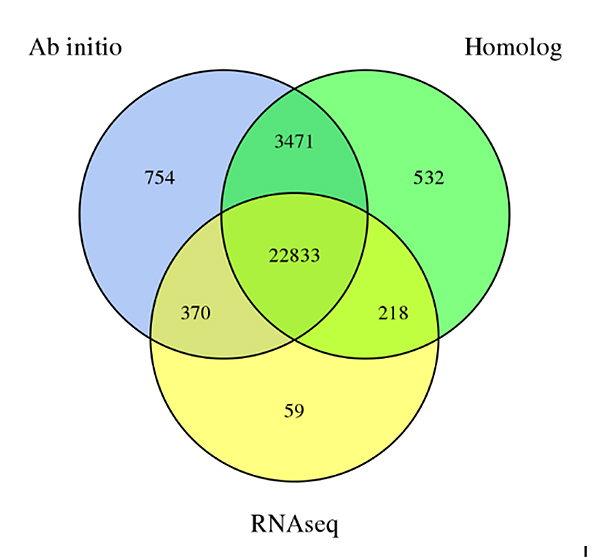
Fu A et al.,Horticulture Research, 2021
4.Kuzivikanwa kweiyo intact refu terminal inodzokorora mumatatu ekotoni genomes
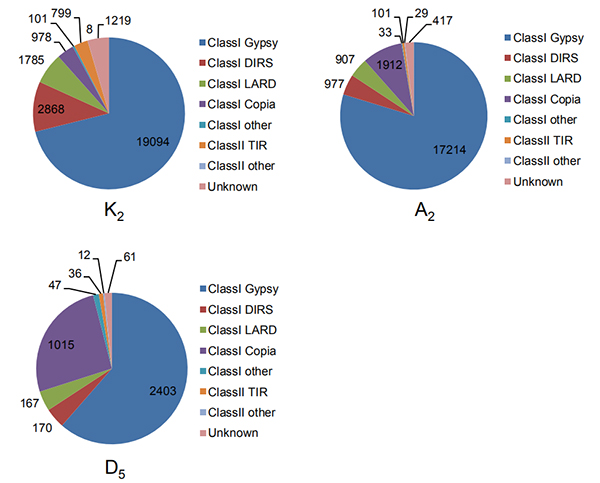
Wang M et al.,Molecular Biology uye Evolution, 2021
5.Hi-C kupisa mepu yeC. acuminatagenome inoratidza genome-wide zvese-ne-zvese kudyidzana.Kusimba kweHi-C kupindirana kwakaenzana kune mutsara kureba pakati pemakontigi.Mutsara wakachena wakatwasuka pamepu yekupisa iyi unoratidza kudzika kwakaringana kwemakontigs pamakromosomes.(Contig anchoring ratio: 96.03%)
kang M et al.,Nature Communications,2021
Nyaya yeBMK
Musangano wepamusoro-soro wegenome unoratidza rye genomic maitiro uye agronomically akakosha majini
Rakatsikiswa: Nature Genetics, 2021
Sequencing strategy:
Genome musangano: PacBio CLR modhi ine 20 kb raibhurari (497 Gb, approx. 63×)
Kugadzirisa kutevedzana: NGS ine 270 bp DNA raibhurari (430 Gb, approx. 54×) pachikuva cheIllumina
Contigs anchoring: Hi-C raibhurari (560 Gb, approx. 71×) paIllumina papuratifomu
Mepu yeOptical: (779.55 Gb, approx. 99×) paBionano Irys
Mibairo yakakosha
1.Musangano weWeining rye genome wakaburitswa uzere genome saizi ye7.74 Gb(98.74% yeinofungidzirwa saizi yegenome nekuyerera kwecytometry).Scaffold N50 yegungano iri yakawana 1.04 Gb.93.67% yemakontigi akabudirira kumiswa pa7 pseudo-chromosomes.Iyi gungano yakaongororwa nemepu yekubatanidza, LAI neBUSCO, izvo zvakakonzera zvibodzwa zvepamusoro muongororo dzese.
2.Kuwedzera zvidzidzo pamusoro pekuenzanisa genomics, genetic linkage mepu, transcriptomics zvidzidzo zvakaitwa pahwaro hwejenome iri.Nhevedzano yehunhu hune hukama hwema genomic maficha akaburitswa anosanganisira genome-wide gene duplications uye maitiro avo pa starch biosynthesis genes;kurongeka kwemuviri kwakaoma prolamin loci, gene expression inoratidzira iri pazasi pekutanga musoro maitiro uye putative domestication-yakabatana chromosomal nzvimbo uye loci mu rye.
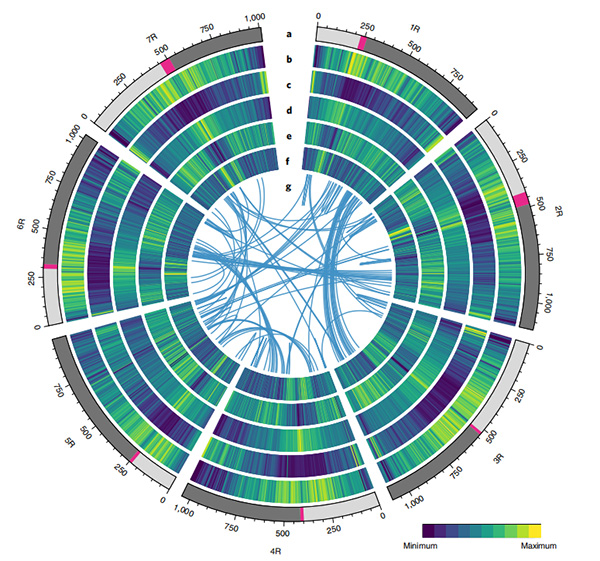 Circos dhizaini pane genomic maficha eWeining rye genome | 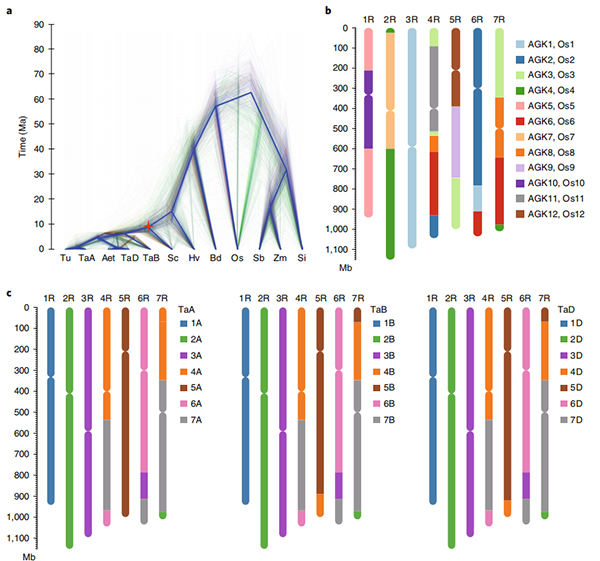 Evolutionary uye chromosome synteny inoongorora ye rye genome |
Li, G., Wang, L., Yang, J.et al.Musangano wepamusoro-soro wegenome unoratidza rye genomic maitiro uye agronomically akakosha majini.Nat Genet 53,574–584 (2021).
https://doi.org/10.1038/s41588-021-00808-z










