T2T GENOME ASSEMBLY, GAP YEMAHARA GENOME
1stMaviri Mupunga Genomes1
Musoro: Gungano uye Kusimbiswa kweMaviri Asina Gap-asina Reference Genomes yeXian/indica Rice Inozivisa Manzwisisiro muPlant Centromere Architecture.
Doi:https://doi.org/10.1101/2020.12.24.424073
Yakatumirwa Nguva: Ndira 01, 2021.
Institute: Huazhong Agricultural University, China
Zvishandiso
O. sativa xian/indicamupunga mhando 'Zhenshan 97 (ZS97)' uye 'Minghui 63 (MH63)
Sequencing strategy
NGS inoverenga + HiFi inoverenga + CLR inoverenga + BioNano + Hi-C
Data:
ZS97: 8.34 Gb (~23x) HiFi inoverenga + 48.39 Gb (~131x ) CLR inoverenga + 25 Gb(~69x) NGS + 2 BioNano Irys masero
MH63: 37.88 Gb (~103x) HiFi inoverenga + 48.97 Gb (~132x) CLR inoverenga + 28 Gb(~76x) NGS + 2 BioNano Irys masero
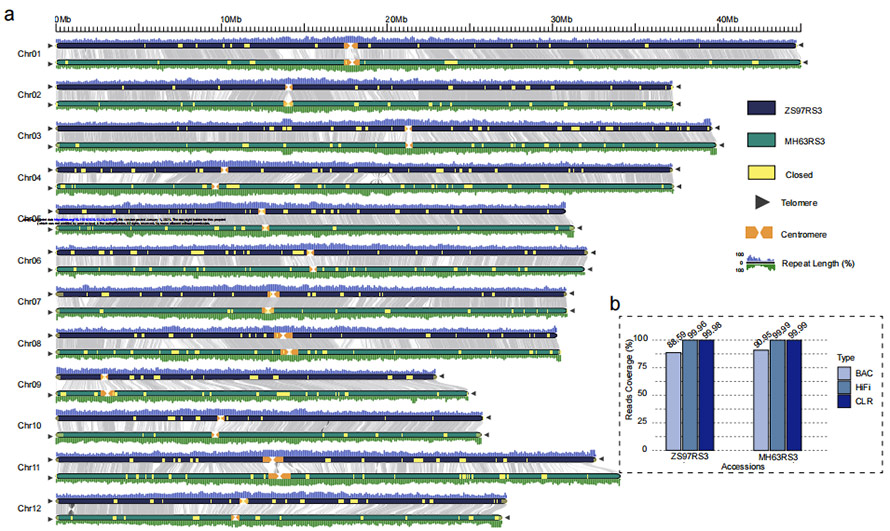
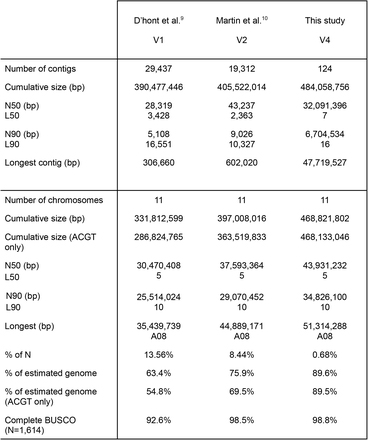
Mufananidzo 1 Maviri-asina gap genomes emupunga (MH63 uye ZS97)
2ndBanana Genome2
Musoro: Telomere-to-telomere gapless chromosome yebhanana inoshandisa nanopore sequencing
Doi:https://doi.org/10.1101/2021.04.16.440017
Yakatumirwa Nguva: Kubvumbi 17, 2021.
Institute: Université Paris-Saclay, France
Zvishandiso
Kaviri haploidMusa acuminatasppmalaccensis(DH-Pahang)
Sequencing strategy uye data:
HiSeq2500 PE250 modhi + MinION/ PromethION (93Gb, ~200X )+ Mepu yeOptical (DLE-1+BspQ1)
Tafura 1 Kuenzanisa kweMusa acuminata (DH-Pahang) genome magungano
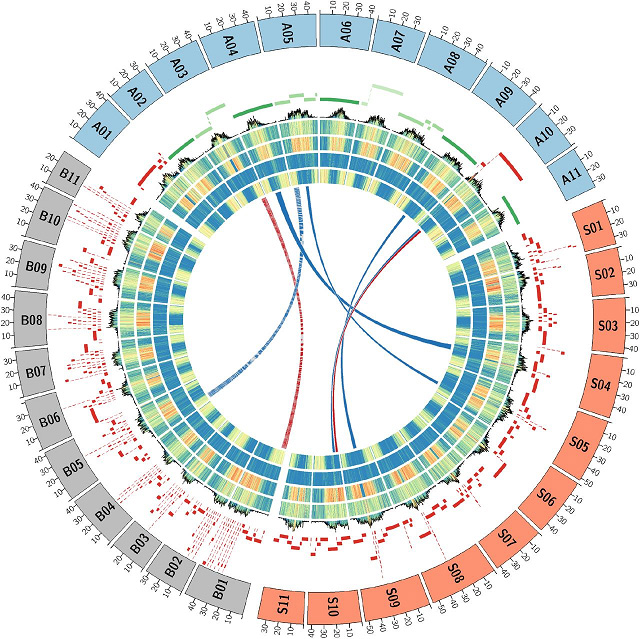
Mufananidzo 2 Musa genomes architecture kuenzanisa
3rdPhaeodactylum tricornutum genome3
Title: Telomere-to-telomere genome assembly yeP
haeodactylum tricornutum
Doi:https://doi.org/10.1101/2021.05.04.442596
Yakatumirwa Nguva: Chivabvu 04, 2021
Sangano: Western University, Canada
Zvishandiso
Phaeodactylum tricornutum(Culture Collection of Algae and Protozoa CCAP 1055/1)
Sequencing strategy uye data:
1 Oxford Nanopore minion inoyerera sero + iyo 2 × 75 yakapetwa-yekupedzisira yepakati-kubuda NextSeq 550 kumhanya
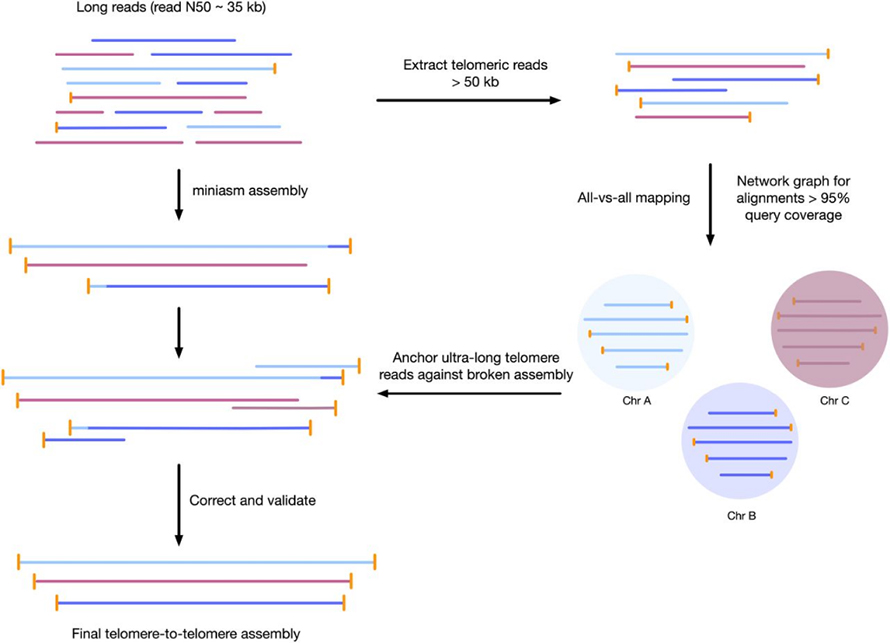
Mufananidzo 3 Kufambiswa kwebasa kwetelomere-to-telomere genome assembly
4thMunhu CHM13 genome4
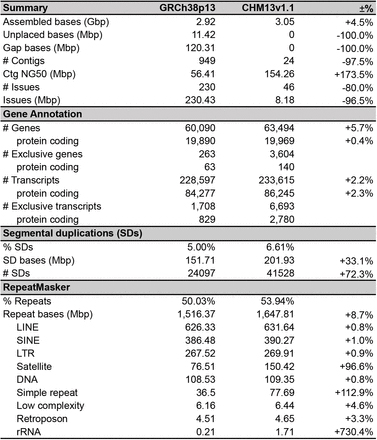
Title: Kutevedzana kwakazara kwegenome remunhu
Doi:https://doi.org/10.1101/2021.05.26.445798
Yakatumirwa Nguva: Chivabvu 27, 2021
Institute: National Institutes of Health (NIH), USA
Zvishandiso: cell line CHM13
Sequencing strategy uye data:
30 × PacBio circular consensus sequencing (HiFi) , 120 × Oxford Nanopore ultra-long kuverenga sequencing , 100 × Illumina PCR-Yemahara sequencing (ILMN) , 70 × Illumina / Arima Genomics Hi-C (Hi-C), BioNano uye Strand-seq
Tafura 2 Kuenzaniswa kweGRCh38 uye T2T-CHM13 magungano evanhu genome
Reference
1.Sergey Nurk et al.Kutevedzana kwakazara kwegenome yemunhu.bioRxiv 2021.05.26.445798;doi:https://doi.org/10.1101/2021.05.26.445798
2.Caroline Belser et al.Telomere-to-telomere gapless chromosomes yebhanana inoshandisa nanopore sequencing.bioRxiv 2021.04.16.440017;doi:https://doi.org/10.1101/2021.04.16.440017
3.Daniel J. Giguere et al.Telomere-to-telomere genome musangano wePhaeodactylum tricornutum.bioRxiv 2021.05.04.442596;doi:https://doi.org/10.1101/2021.05.04.442596
4.Jia-Ming Song et al.Gungano uye Kusimbiswa kweMaviri Asina Gap-asina Reference Genomes yeXian/indica Rice Inozivisa Manzwisisiro muPlant Centromere Architecture.bioRxiv 2020.12.24.424073;doi:https://doi.org/10.1101/2020.12.24.424073
Nguva yekutumira: Jan-06-2022

