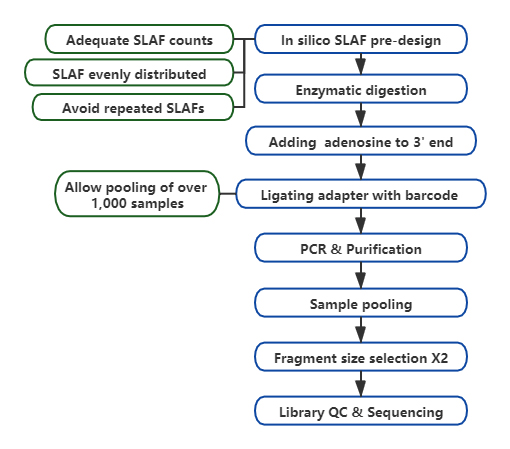
High-throughput genotyping, kunyanya pahuwandu hwevanhu, inhanho inokosha muzvidzidzo zvekubatana kwemajini, izvo zvinopa genetic nheyo yekushanda kwemajini ekutsvaga, evolutionary analysis, etc. Panzvimbo yekudzika kwese genome re-sequencing, kuderedza kumiririra genome sequencing (RRGS). ) inounzwa kuti ideredze mutengo wekuteedzana pasample, uku uchichengetedza kugona kwakanaka pakuwana genetic marker.Izvi zvinowanzo wanikwa nekubvisa chimedu chezvirambidzo mukati meiyo saizi yakapihwa renji, iyo inonzi yakaderedzwa yekumiririra raibhurari (RRL).Specific-locus amplified fragment sequencing(SLAF-Seq) inzira yekuzvigadzira yega ye de novo SNP kuwanikwa uye SNP genotyping yehuwandu hwevanhu.
Technical workflow
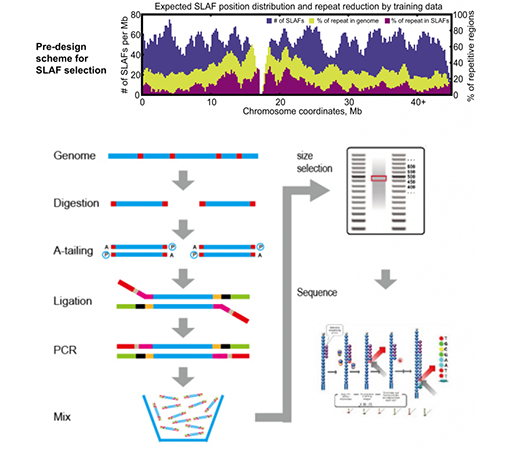
SLAF vs iripo RRL nzira
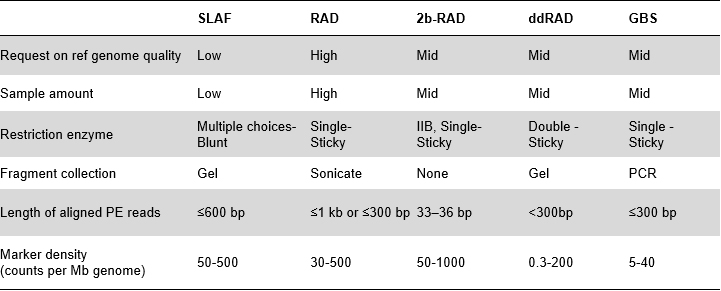
Zvakanakira zveSLAF
Yepamusoro genetic marker yekuwana kugona-Yakasanganiswa nehunyanzvi hwekutevedzanisa tekinoroji, SLAF-Seq inogona kuwana mazana ezviuru ematagi akawanikwa mukati megenome yese kuzadzisa chikumbiro chezvirongwa zvekutsvagisa zvakasiyana, kungave kana nereferensi genome.
Customized & Flexible kuedza dhizaini-Kune chinangwa chekutsvagisa chakasiyana kana mhando, nzira dzakasiyana dzekudyara enzymatic dziripo dzinosanganisira single-enzyme, dual-enzyme uye akawanda-enzyme digestion.Digestion strategy ichafanoongororwa musilico kuvimbisa yakakwana enzyme dhizaini.
Kushanda kwakanyanya mu enzymatic digestion- Pre-yakagadzirwa enzymatic digestion inopa zvakanyanya kugoverwa maSLAF pachromosome.Zvimedu zvekuunganidza zvinobudirira zvinogona kuwana pamusoro pe95%.
Dzivisa kudzokorora kutevedzana- Mazana ekudzokororwa kutevedzana muSLAF-Seq data yakaderedzwa kusvika yakaderera kupfuura 5%, kunyanya mumhando dzine huwandu hwepamusoro hwezvinhu zvinodzokororwa, segorosi, chibage, nezvimwe.
Kuzvigadzira pachako bioinformamatic workflow- BMK yakagadzira yakasanganiswa bioinformamatic workflow inoshanda kune SLAF-Seq tekinoroji kuve nechokwadi chekuvimbika uye nekurongeka kwekubuda kwekupedzisira.
Kushandiswa kweSLAF
Genetic linkage mepu
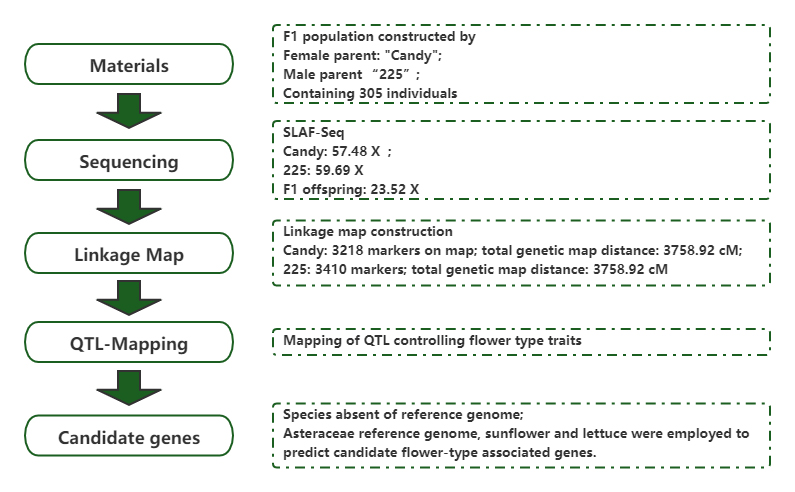
High-density genetic mepu kuvaka uye kuzivikanwa kweloci inodzora maruva-mhando hunhu muChrysanthemum(Chrysanthemum x morifolium Ramat.)
Chinyorwa: Tsvakurudzo yeHorticulture Yakabudiswa: 2020.7
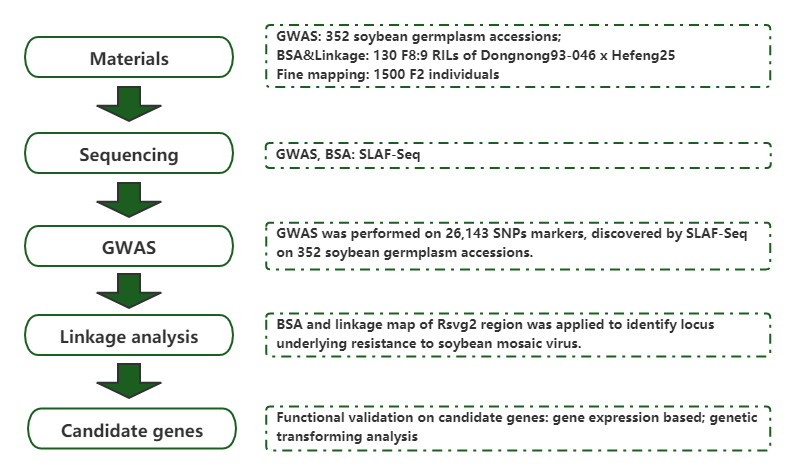
GWAS
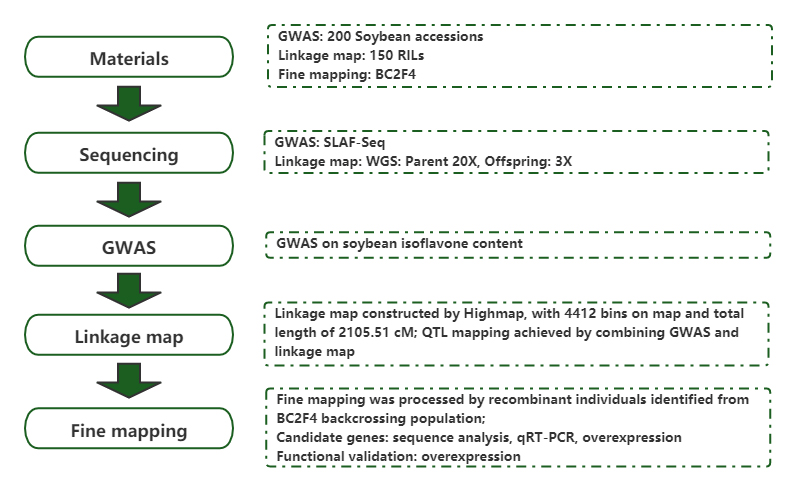
Kuzivikanwa kwejini remumiriri rakabatana neisofavone zvirimo mumhodzi dzesoya uchishandisa genome-wide mubatanidzwa uye yekubatanidza mepu.
Nyaya: The Plant Journal Yakabudiswa: 2020.08
Evolutionary Genetics
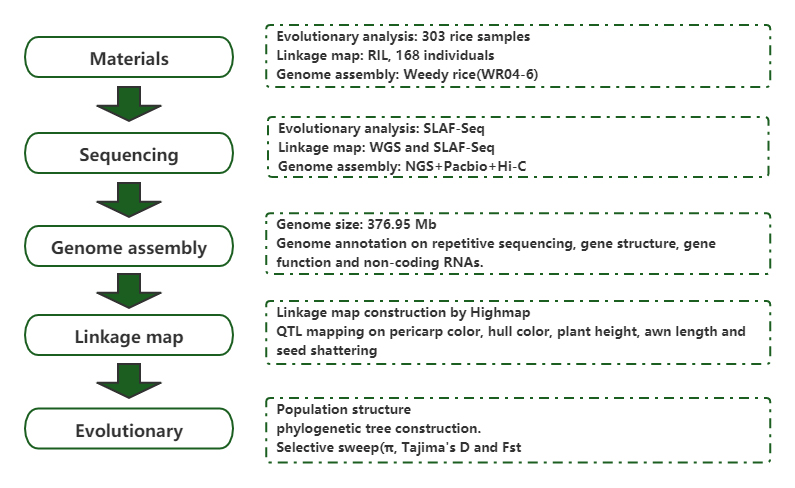
Population genomic analysis uye de novo assembly inoratidza kwakabva sora remupunga semutambo wekushanduka
Chinyorwa: Molecular Plant Yakabudiswa: 2019.5
Bulked Segregant Analysis (BSA)
GmST1, iyo inoisa sulfotransferase, inopa kuramba kune soyabean mosaic virus strains G2 uye G3.
Chinyorwa: Chidyarwa, Sero & Nezvakatipoteredza Rakabudiswa: 2021.04
Reference
Zuva X, Liu D, Zhang X, nevamwe.SLAF-Seq: nzira inoshanda yehombe-scale de novo SNP kuwanikwa uye genotyping uchishandisa yakakwirira-kuburikidza sequencing[J].Plos one, 2013, 8(3):e58700
Rwiyo X, Xu Y, Gao K, nevamwe.High-density genetic mepu kuvaka uye kuzivikanwa kweloci inodzora maruva-mhando hunhu muChrysanthemum (Chrysanthemum × morifolium Ramat.).Hortic Res.2020;7:108.
Wu D, Li D, Zhao X, nevamwe.Kuzivikanwa kwejini rine hukama neisoflavone zvirimo mumhodzi dzesoya uchishandisa genome-wide mubatanidzwa uye yekubatanidza mepu.Plant J. 2020;104(4): 950-963.
Sun J, Ma D, Tang L, nevamwe.Population Genomic Analysis uye De Novo Assembly Inoburitsa Mabviro eWeedy Rice seMutambo Wekushanduka.Mol Plant.2019;12(5):632-647.Mol Plant.2018;11(11):1360-1376.
Zhao X, Jing Y, Luo Z, nevamwe.GmST1, iyo inoisa sulfotransferase, inopa kuramba kune soyabean mosaic virus strains G2 neG3.Plant Cell Environ.2021;10.1111/pce.14066
Nguva yekutumira: Jan-04-2022