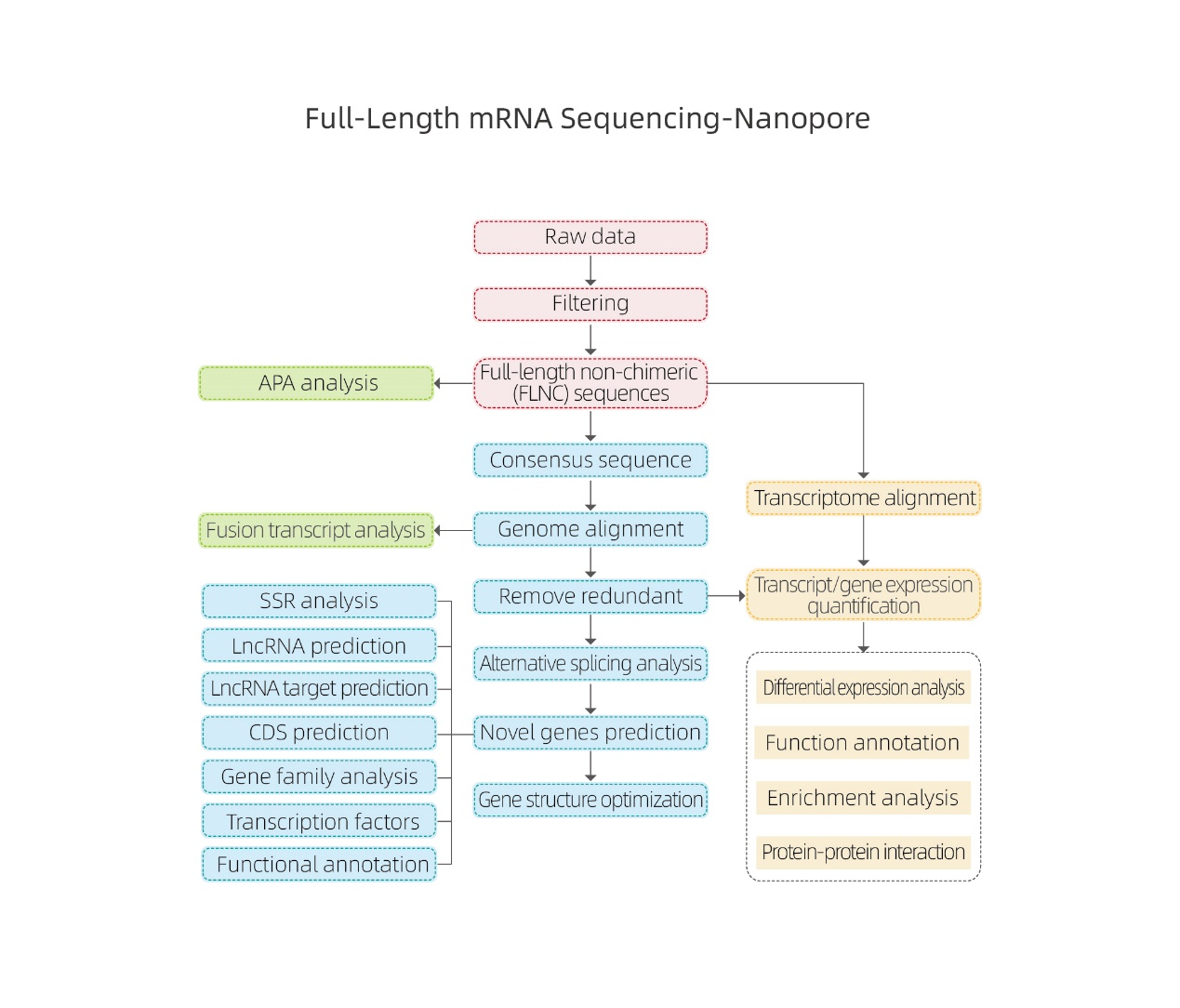
Yakazara-Kureba mRNA Sequencing-Nanopore
Basa Rinobatsira
● Kusarura kwekutevedzana kwakaderera
● Kuburitsa mamorekuru ecDNA akareba
● data shoma inodiwa kuvhara nhamba imwechete yezvinyorwa
● Kuzivikanwa kweakawanda isoforms per gene
● Kuwedzera kwekutaura mune isoform level
Sevhisi Zvinotsanangurwa
| Library | Platform | Inokurudzirwa kuburitsa data (Gb) | Quality Control |
| cDNA-PCR(Poly-A yakafumiswa) | Nanopore PromethION P48 | 6 Gb/sample (Zvichienderana nemhando) | Hurefu hwakazara chiyero>70% Avhareji yemhando yepamusoro: Q10
|
Bioinformatics inoongorora
●Raw data processing
● Kuzivikanwa kwezvinyorwa
● Alternative splicing
● Kuwedzera kwekutaura mune gene level uye isoform level
● Kuongorora kwekutaura kwakasiyana
● Basa rekuzivisa uye kupfumisa (DEGs neDETs)
Sample Zvinodiwa uye Delivery
Sample Zvinodiwa:
Nucleotides:
| Conc.(ng/μl) | Mari (μg) | Kuchena | Kuperera |
| ≥ 100 | ≥ 0.6 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Yakaganhurirwa kana isina mapuroteni kana kusvibiswa kweDNA kunoratidzwa pagel. | Kumiti: RIN≥7.0; Kumhuka: RIN≥7.5; 5.0≥28S/18S≥1.0; kukwirira kwakadzika kana kusina kwekutanga |
Matumbu: Huremu(yakaoma): ≥1 g
*Kune tishu diki pane 5 mg, tinokurudzira kutumira flash yakaomeswa nechando (mumvura nitrogen) sampuli yetishu.
Kumiswa kwesero: Kuwanda kwesero = 3 × 1061 × 107
* Isu tinokurudzira kutumira chando cell lysate.Kana iyo sero inoverenga idiki pane 5 × 105, flash yakaomeswa mu nitrogen yemvura inokurudzirwa, iyo inosarudzika kune micro extraction.
Mienzaniso yeropa: Vhoriyamu≥1 ml
Yakakurudzirwa Sample Delivery
Container: 2 ml centrifuge chubhu (Tin foil haina kukurudzirwa)
Sample labeling: Boka+ replicate eg A1, A2, A3;B1, B2, B3...
Shipment: 2, Dry-ice: Samples dzinoda kutakurwa mumabhegi uye kuvigwa muoma-aizi.
- RNAstable chubhu: RNA samples inogona kuomeswa muRNA kudzikamisa chubhu (eg RNAstable®) uye kutumirwa mukamuri tembiricha.
Kuyerera Kwebasa Rebasa
Nucleotides:

Muenzaniso wekutumira

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
Kuyerera Kwebasa Rebasa
Tissue:

Kuedza kugadzira

Muenzaniso wekutumira

RNA kubviswa

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
1.Differential expression analysis -Volcano plot
Kuongorora kwakasiyana kwekutaura kunogona kugadziridzwa mune ese gene nhanho kuti aone akasiyana anoratidzwa majini (DEGs) uye mune isoform level kuona zvakasiyana.
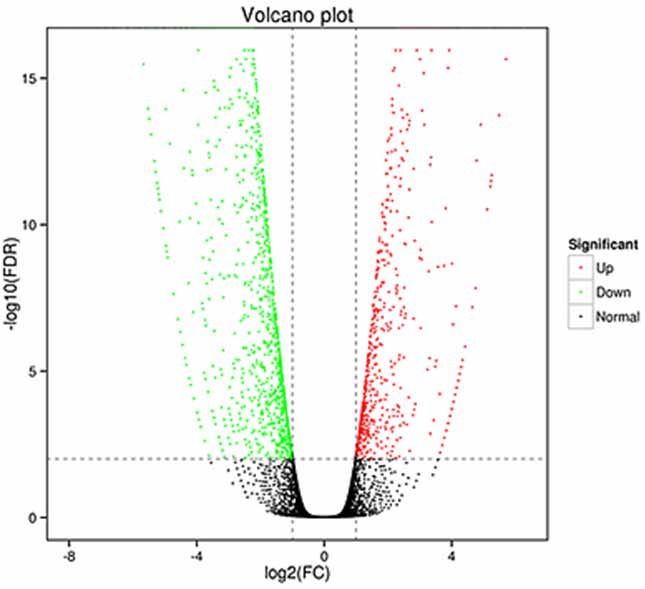
zvakanyorwa zvinyorwa (DETs)
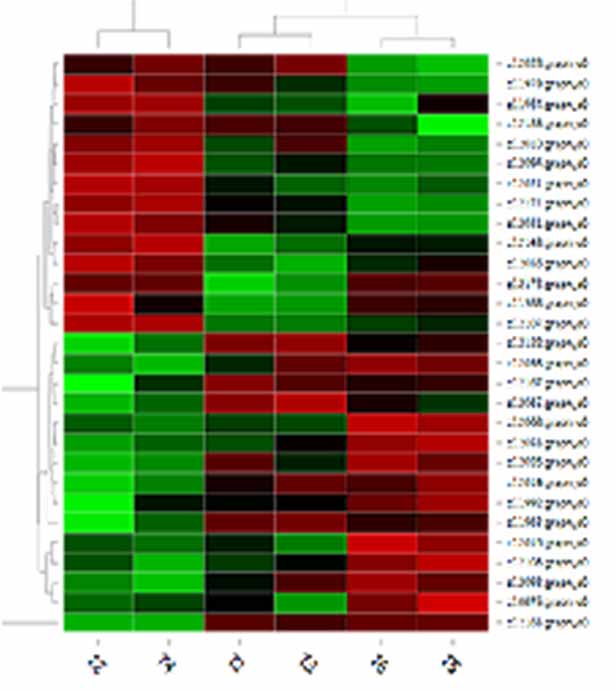
2.Hierarchical clustering heatmap
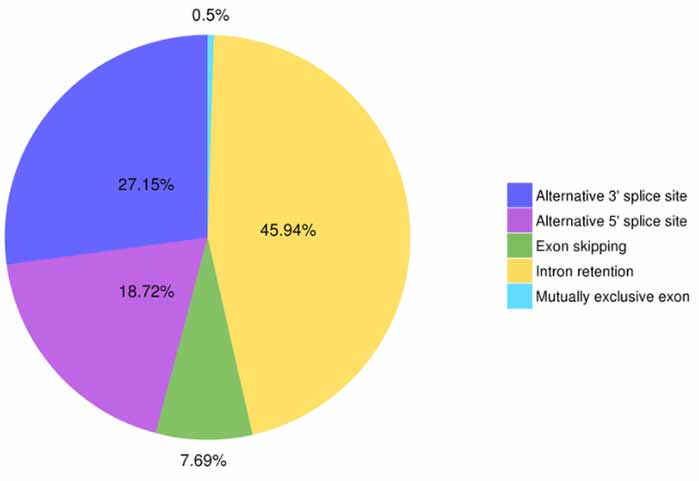
3.Alternative splicing identification uye classification
Mhando shanu dzezvimwe zviitiko splicing zvinogona kufanotaurwa neAstalavista.
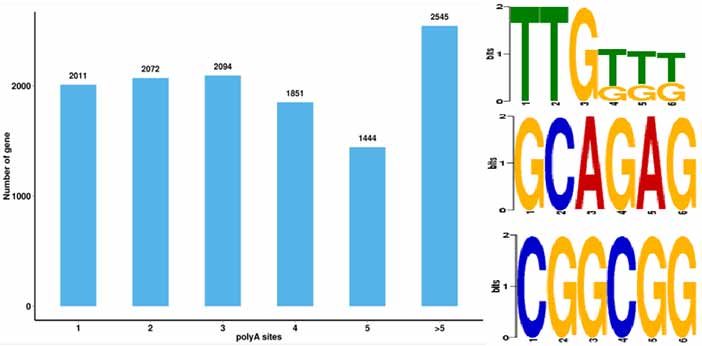
4.Alternative poly-adenylation (APA) zviitiko zvekuzivikanwa uye Motif pa50 bp kumusoro kwepoly-A
Nyaya yeBMK
Alternative splicing identification uye isoform-level quantification ne nanopore full-length transcriptome sequencing.
Rakatsikiswa:Nature Communications, 2020
Sequencing strategy:
Kuunganidza: 1. CLL-SF3B1(WT);2. CLL-SF3B1(K700E mutation);3. Kazhinji B-masero
Sequencing zano: MinION 2D raibhurari sequencing, PromethION 1D raibhurari sequencing;data pfupi-yekuverenga kubva kumasampuli akafanana
Sequencing platform: Nanopore MinION;Nanopore PromethION;
Mibairo yakakosha
1.Isoform-level Alternative Splicing Identification
Kuverengera kwenguva refu kutevedzana kunopa simba kuzivikanwa kwemutant SF3B1K700E-altered splice sites at isoform-level.35 mamwe ma3′SS uye gumi mamwe ma5′SS akawanikwa akapatsanurwa zvakanyanya pakati peSF3B1.K700Euye SF3b1WT.33 ye 35 shanduko dzaive dzichangowanikwa nekuverenga kwenguva refu kutevedzana.
2.Isoform-level Alternative Splicing quantification
Tsanangudzo ye intron retention (IR) isoforms muSF3B1K700Euye SF3b1WTakaverengerwa zvichienderana nenanopore sequences, zvichiratidza pasi-pasi-mutemo weIR isoforms muSF3B1.K700E.
Reference
Tang AD , Soulette CM , Baren MJV , et al.Yakazara-yakareba-yakanyorwa maratidziro eSF3B1 mutation mune isingaperi lymphocytic leukemia inoratidza kuderedzwa kweakachengetwa introns [J].Nature Communications.