
Evolutionary Genetics
Basa Rinobatsira
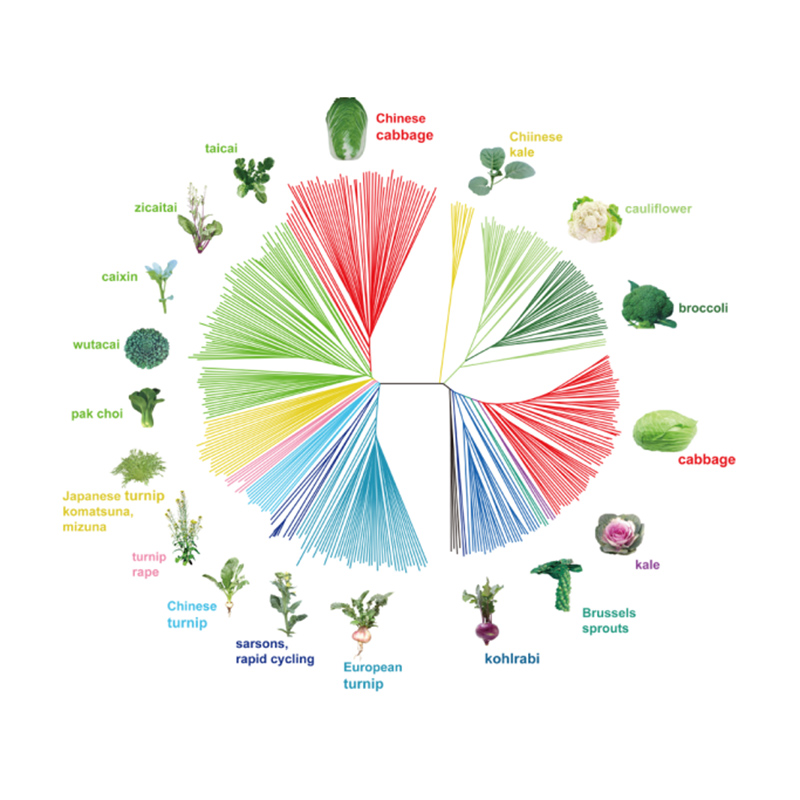
Takagi et al.,The plant journal, 2013
● Kufungidzira kusiyana kwemarudzi nguva nekumhanya kunoenderana nekusiyana kwema nucleotide nemaamino acids.
● Kuburitsa hukama hwakavimbika hwephylogenetic pakati pezvipenyu zvine simba shoma rekushanduka-shanduka nekushanduka-shanduka
● Kugadzira hukama pakati pekuchinja kwemajini uye phenotypes kufumura maitiro ane hukama
● Kufungidzira kusiyana-siyana kwemavara okugara nhaka, kunoratidza kushanduka-shanduka kwemarudzi
● Kukurumidza Kuchinja Nguva
● Zvakawanda ruzivo: BMK yakaunganidza ruzivo rwakakura muhuwandu hwevanhu uye mapurojekiti ane chekuita nekushanduka-shanduka kwemakore anopfuura gumi nemaviri, achifukidza mazana emarudzi, nezvimwewo uye akabatsira mumapurojekiti anopfuura makumi masere epamusoro-soro anoburitswa muNature Communications, Molecular Plants, Plant Biotechnology Journal, nezvimwewo.
Sevhisi Zvinotsanangurwa
Zvekushandisa:
Kazhinji, vanhu vanosvika vatatu (eg subspecies kana strains) vanokurudzirwa.Huwandu hudiki hwega hwega hunofanirwa kuve nevanhu vasingasviki gumi (Zvirimwa >15, zvinogona kudzikiswa kune zvisingawanzo zvisikwa).
Sequencing strategy:
* WGS inogona kushandiswa kumarudzi ane emhando yepamusoro referensi genome, ukuwo SLAF-Seq inoshanda kune zvipenyu zvine kana zvisina referensi genome, kana referensi genome yehurombo.
| Inoshanda kune genome size | WGS | SLAF-Tags (×10,000) |
| ≤ 500 Mb | 10×/munhu mumwe nomumwe | WGS inokurudzirwa zvakanyanya |
| 500 Mb - 1 Gb | 10 | |
| 1 Gb - 2 Gb | 20 | |
| ≥2 Gb | 30 |
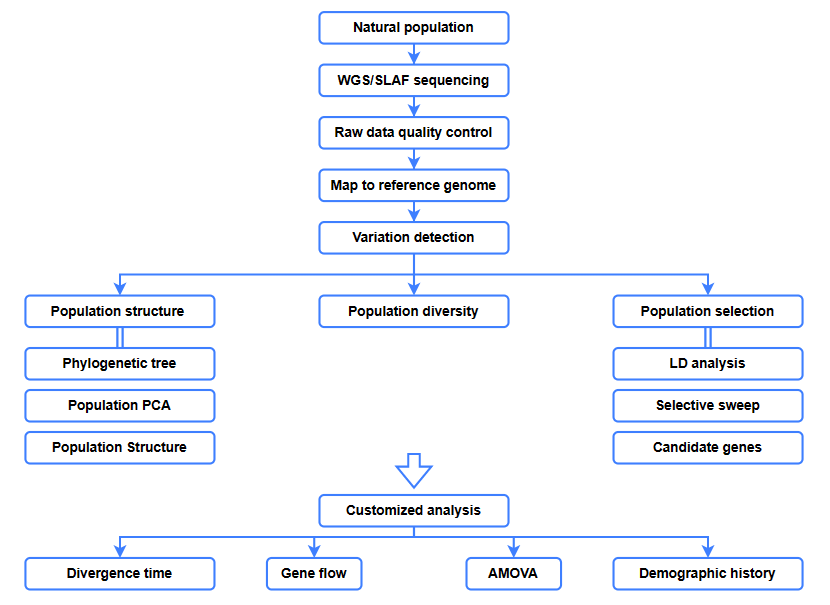
Bioinformatics inoongorora
● Evolutionary analysis
● Kutsvaira kunosarudza
● Kuyerera kwemajini
● Nhoroondo yevanhu
● Nguva yekusiyana
Sample Zvinodiwa uye Delivery
Sample Zvinodiwa:
| Zvisikwa | Tissue | WGS-NGS | SLAF |
| Mhuka
| Visceral tishu |
0.5~1g
|
0.5g
|
| Tsandanyama | |||
| Ropa reMammalian | 1.5mL
| 1.5mL
| |
| Huku/Hove ropa | |||
| Plant
| Fresh Leaf | 1~2g | 0.5~1g |
| Petal/Stem | |||
| Mudzi/Mbeu | |||
| Masero | Cultured sero |
| gDNA | Concentration | Mari (ug) | OD260/OD280 |
| SLAF | ≥35 | ≥1.6 | 1.6-2.5 |
| WGS-NGS | ≥1 | ≥0.1 | - |
Kuyerera Kwebasa Rebasa

Kuedza kugadzira

Muenzaniso wekutumira

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
*Demo mhedzisiro inoratidzwa pano yese kubva kugenomes yakaburitswa neBMKGENE
1.Evolution analysis ine kuvakwa kwe phylogenetic tree, chimiro chevanhu uye PCA inobva pane zvakasiyana-siyana.
Phylogenetic muti inomiririra taxonomic uye hukama hwekushanduka-shanduka pakati pemarudzi ane madzitateguru akafanana.
PCA ine chinangwa chekuona padhuze pakati pehuwandu hwevanhu.
Population chimiro chinoratidza kuvepo kweiyo genetically distinct sub-population maererano neallele frequencies.
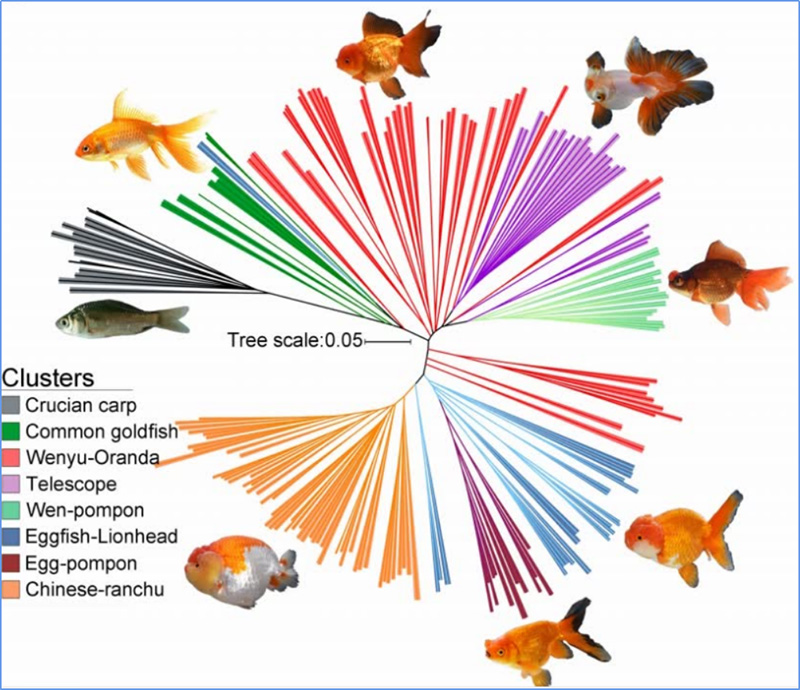
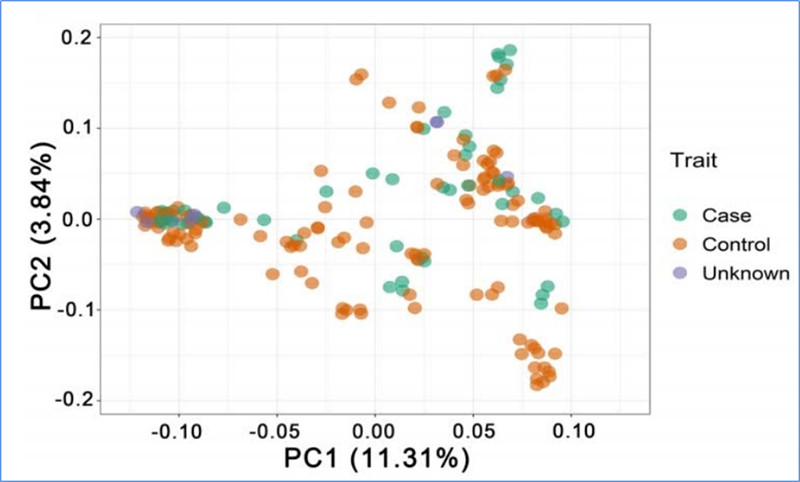
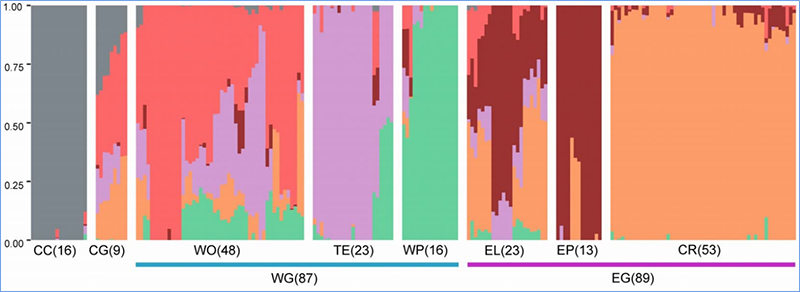
Chen, uye.al.,PNAS, 2020
2.Selective tsvaira
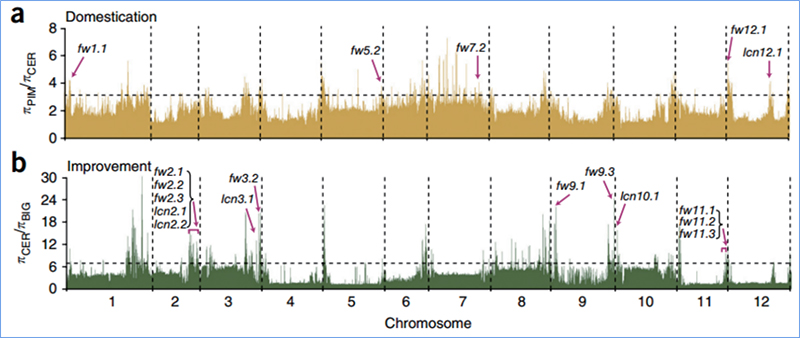
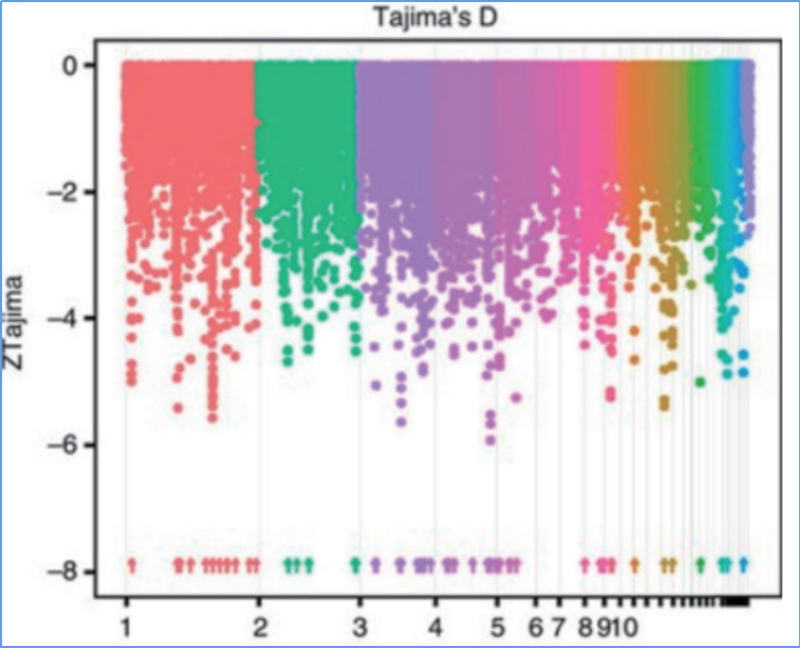
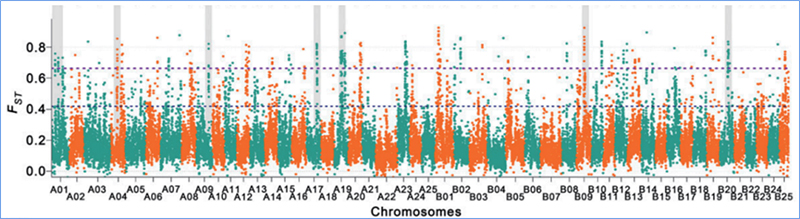
Sarudzo yekutsvaira inoreva nzira inosarudzwa saiti inobatsira uye mafambiro enzvimbo dzakabatana asina kwaakarerekera anowedzerwa uye ayo enzvimbo dzisina kubatana anodzikiswa, zvichikonzera kudzikiswa kwedunhu.
Kuonekwa kweGenome-wide pamatunhu akasarudzika anogadziriswa nekuverenga huwandu hwevanhu genetic index (π,Fst, Tajima's D) yeese maSNP mukati mehwindo rinotsvedza (100 Kb) pane imwe nhanho (10 KB).
Nucleotide kusiyana (π)
Tajima's D
Fixation index (Fst)
Wu, uye.al.,Molecular Plant, 2018
3.Gene Kuyerera
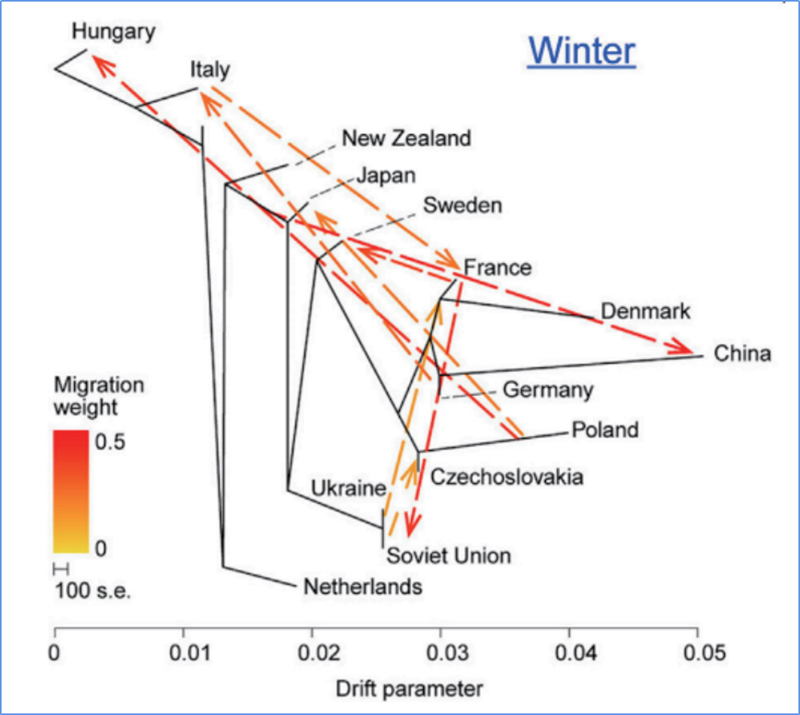
Wu, uye.al.,Molecular Plant, 2018
4.Demographic nhoroondo
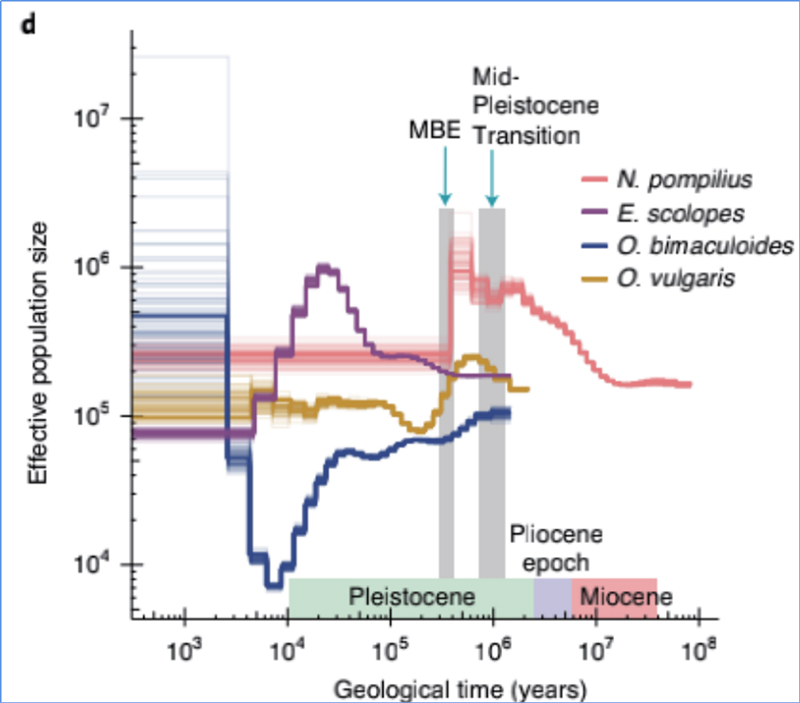
Zhang, nevamwe.al.,Nature Ecology & Evolution, 2021
5.Divergence nguva
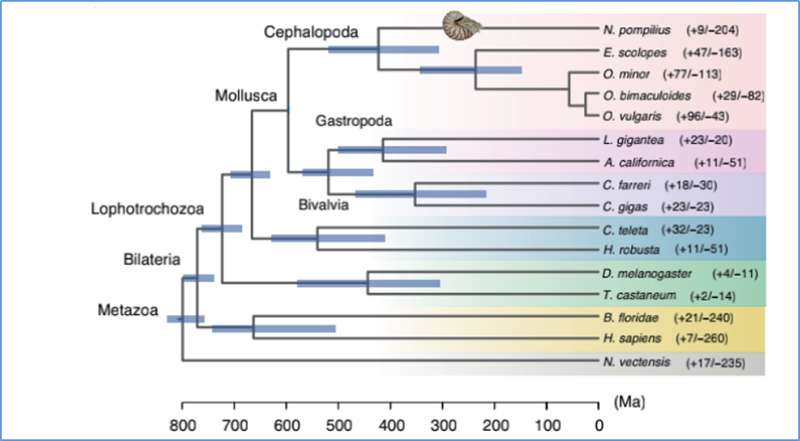
Zhang, nevamwe.al.,Nature Ecology & Evolution, 2021
Nyaya yeBMK
Mepu ye genomic variation inopa nzwisiso muhwaro hwemajini eSpring Chinese Cabbage(Brassica rapa ssp. Pekinensis) sarudzo.
Rakatsikiswa: Molecular Plant, 2018
Sequencing strategy:
Resequencing: kutevedzana kudzika: 10 ×
Mibairo yakakosha
Muchidzidzo ichi, 194 Chinese makabheji akagadziriswa kuti aenzaniswe zvakare neavhareji kudzika kwe10×, iyo yakapa 1,208,499 SNPs uye 416,070 InDels.Kuongorora kwePhylogenetic pamitsara iyi ye194 yakaratidza kuti mitsara iyi inogona kugoverwa kuva ecotypes matatu, chitubu, zhizha uye matsutso.Pamusoro pezvo, chimiro chehuwandu uye kuongororwa kwePCA kwakaratidza kuti chitubu Chinese cabbage yakatanga kubva kumatsutso makabheji muShandong, China.Izvi zvakazounzwa kuKorea neJapan, zvakayambuka nemitsetse yemuno uye mamwe marudzi ekunonoka-bolting awo akadzoserwa kuChina uye akazova Spring Chinese cabbage.
Genome-wide scanning pamatsutso eChinese makabheji uye mumatsutso makabheji pakusarudza kwakaratidza 23 genomic loci dzakapfuura nekusarudzwa kwakasimba, maviri acho aipindirana nenzvimbo yekudzora-nguva yekutonga zvichienderana neQTL-mepu.Matunhu maviri aya akawanikwa aine majini akakosha anodzora kutumbuka, BrVIN3.1 uye BrFLC1.Aya majini maviri akasimbiswa zvakare kuve akabatanidzwa mukuvhara nguva nekudzidza kwezvinyorwa uye transgenic kuyedza.
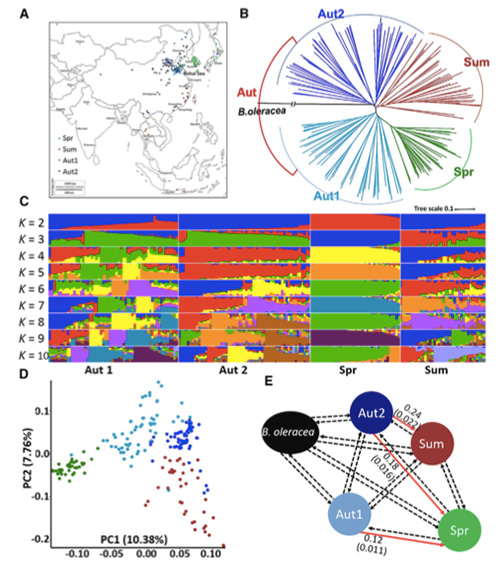 Population structure analysis paChinese cabbage | 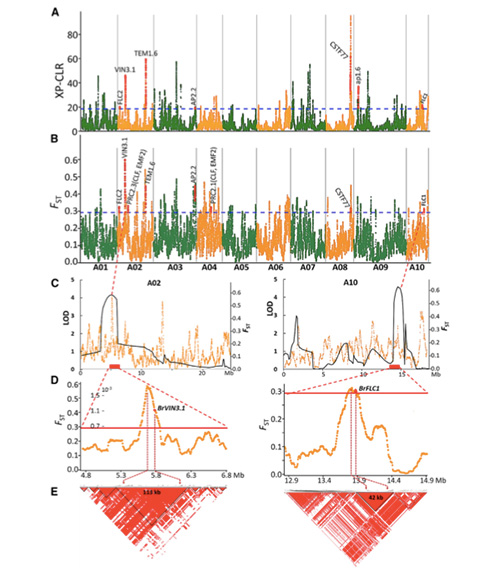 Ruzivo rweGenetic pakusarudza kweChinese kabichi |
Tongbing, et al."A Genomic Variation Mepu Inopa Insights muGenetic Basis yeChitubu Chinese Cabbage (Brassica rapa ssp.pekinensis)Sarudzo."Molecular Plants,11(2018):1360-1376.















