
Bulked Segregant analysis
Basa Rinobatsira
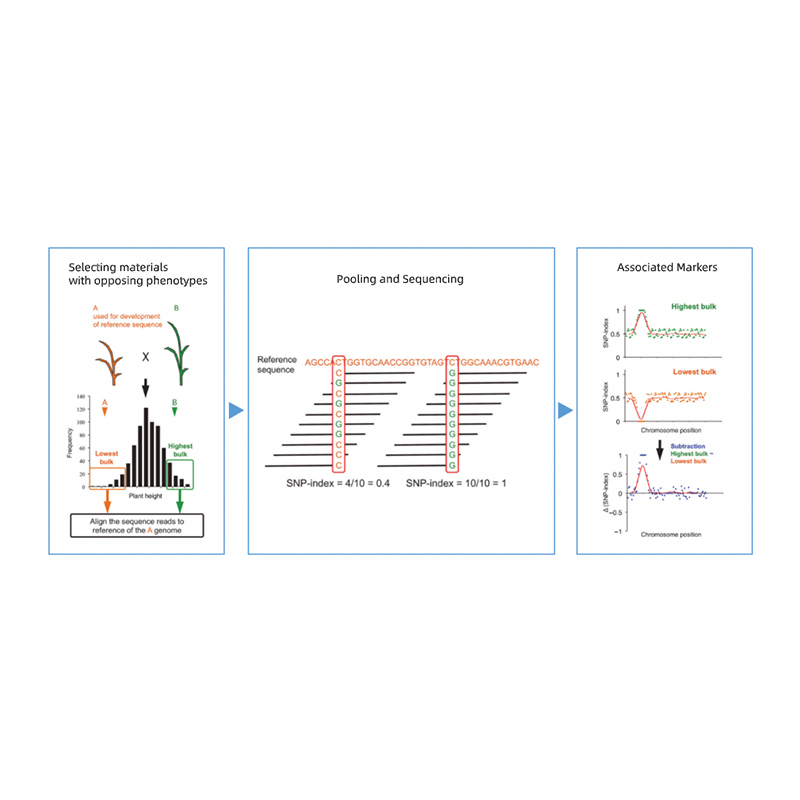
Takagi et al., The plant Journal, 2013
● Yakarurama yenzvimbo: Kusanganisa hukuru ne30 + 30 kusvika ku200 + 200 vanhu kuti kuderedza ruzha rwemashure;vasiri-synonymous mutatants-based candidate dunhu kufanotaura.
● Kuongorora kwakadzama: In-depth candidate gene function annotation, kusanganisira NR, SwissProt, GO, KEGG, COG, KOG, nezvimwewo.
● Kurumidza Kuchinja Nguva: Kurumidza kugadzirwa kwegene mukati me45 mazuva ekushanda.
● Ruzivo rwakadzama: BMK yakabatsira muzviuru zvehunhu hwekugara, hunosanganisira marudzi akasiyana-siyana sezvirimwa, zvigadzirwa zvemumvura, sango, maruva, michero, nezvimwe.
Sevhisi Zvinotsanangurwa
Population:
Kuparadzanisa vana vevabereki vane phenotypes inopikisa.
semuenzaniso F2 chizvarwa, Backcrossing (BC), Recombinant inbred line (RIL)
Kusanganisa dziva
Kune hunhu hwemhando: 30 kune 50 vanhu (zvishoma makumi maviri) / zvakawanda
Pahuwandu hwetratis: pamusoro 5% kusvika 10% vanhu vane phenotypes yakanyanyisa muhuwandu hwese (zvishoma 30 + 30).
Inokurudzirwa kutevedzana kudzika
Kanenge 20X/mubereki uye 1X/mwana munhu (semuenzaniso kuvana kusanganisa dziva re30+30 munhu, kutevedzana kudzika kuchave 30X pahuwandu)
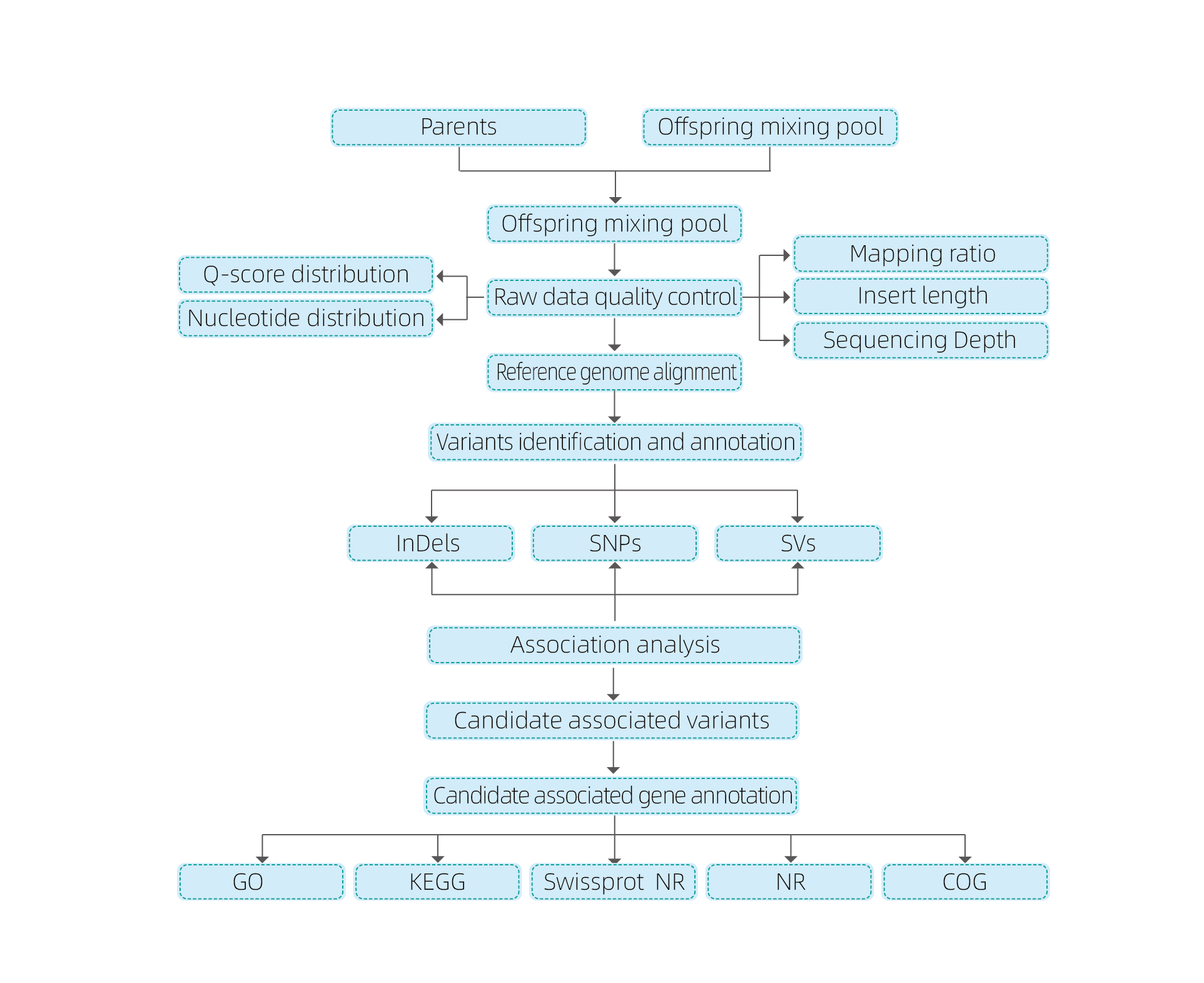
Bioinformatics inoongorora
● Genome resequencing
● Kugadziriswa kwemashoko
● SNP/Indel kufona
● Kuongororwa kwedunhu remumiriri
● Candidate gene function annotation
Sample Zvinodiwa uye Delivery
Sample Zvinodiwa:
Nucleotides:
| gDNA muenzaniso | Muenzaniso wenyama |
| Kuisa pfungwa: ≥30 ng/μl | Zvirimwa: 1-2 g |
| Kuwanda: ≥2 μg (Vhoriyamu ≥15 μl) | Mhuka: 0.5-1 g |
| Kuchena: OD260/280= 1.6-2.5 | Ropa rose: 1.5 ml |
Kuyerera Kwebasa Rebasa

Kuedza kugadzira

Muenzaniso wekutumira

RNA kubviswa

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
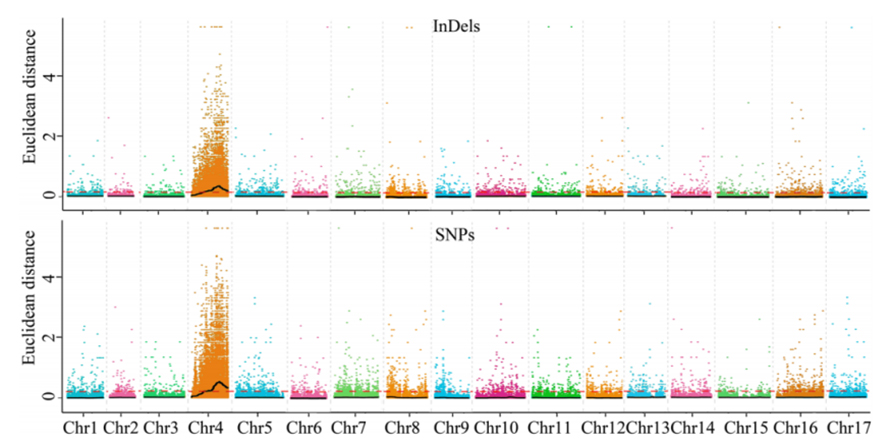
1.Association analysis base paEuclidean Distance (ED) kuti uone dunhu remumiriri.Mumufananidzo unotevera
X-axis: Chromosome nhamba;Doti rega rega rinomiririra kukosha kweED kweSNP.Mutsetse weBlack unoenderana neyakakwana ED kukosha.Iyo yepamusoro ED kukosha inoratidza kushamwaridzana kwakakosha pakati pesaiti uye phenotype.Red dash line inomiririra chikumbaridzo chekubatana kwakakosha.
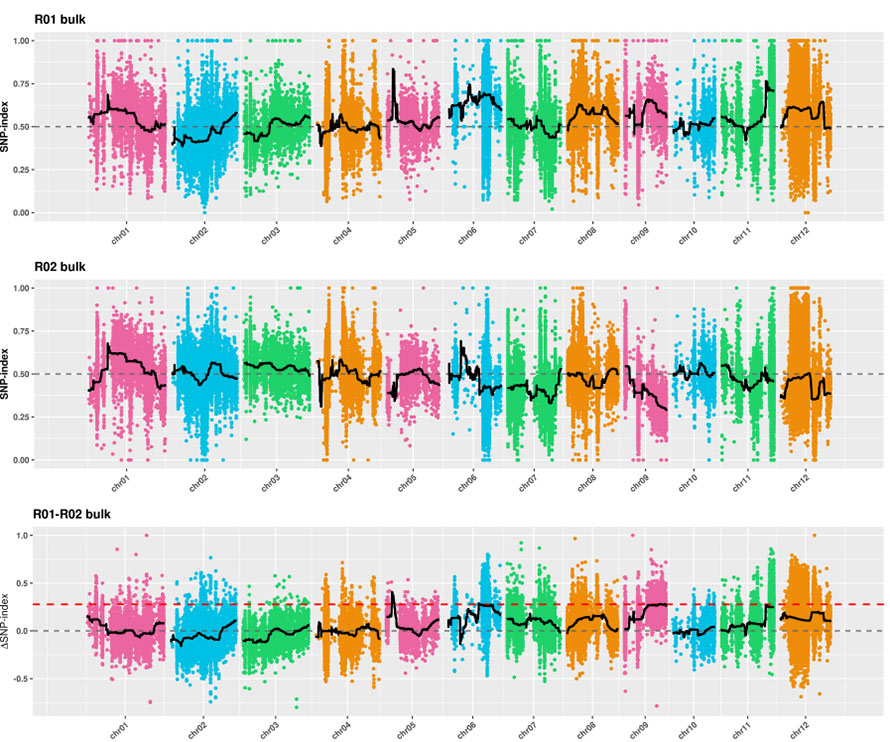
2.Association analysis based no SNP-index
X-axis: Chromosome nhamba;Imwe neimwe dot inomiririra SNP-index kukosha.Mutsetse mutema unomirira kuti fitted SNP-index value.Iyo yakakura kukosha, iyo inonyanya kukosha kushamwaridzana.
Nyaya yeBMK
Iyo huru-effect quantitative trait locus Fnl7.1 inovharira kunonoka embryogenesis abundant protein ine chekuita nehureba hwemutsipa mucucumber.
Rakatsikiswa: Plant Biotechnology Journal, 2020
Sequencing strategy:
Vabereki (Jin5-508, YN): Yese genome resequencing ye34× uye 20×.
DNA madziva (50 Yakareba-mitsipa uye 50 mitsipa mipfupi): Resequencing ye61× uye 52×
Mibairo yakakosha
Muchidzidzo ichi, kupatsanura vanhu (F2 neF2: 3) kwakagadzirwa nekuyambuka refu-neck cucumber line Jin5-508 uye pfupi-mutsipa YN.Madziva maviri eDNA akavakwa ne50 yakanyanyisa mitsipa mirefu uye 50 yakanyanyisa-pfupi-mitsipa vanhu.Huru-effect QTL yakaonekwa paChr07 neBSA ongororo uye yechinyakare QTL mepu.Dunhu remumiriri rakadzikiswa zvakare nekuita mepu yakanaka, gene expression quantification uye transgenic kuyedza, iyo yakaburitsa kiyi geni mukudzora huro-hurefu, CsFnl7.1.Mukuwedzera, polymorphism muCsFnl7.1 inosimudzira nharaunda yakawanikwa ichibatanidzwa neinofanana kutaura.Imwezve phylogenetic ongororo yakaratidza kuti Fnl7.1 locus inogona kunge yakabva kuIndia.
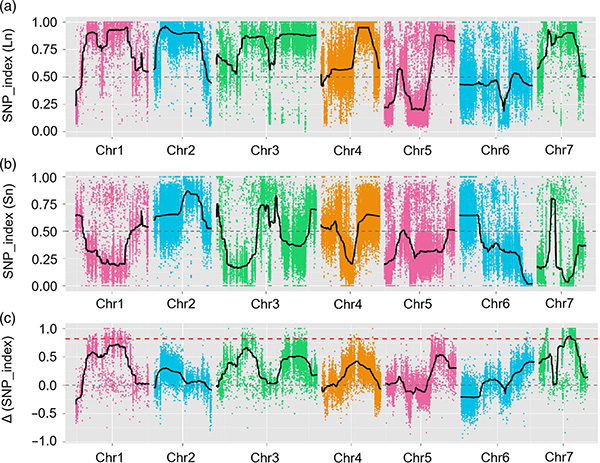 QTL-mapping muBSA ongororo yekuona dunhu remumiriri rakabatana nehurefu hwehuro cucumber | 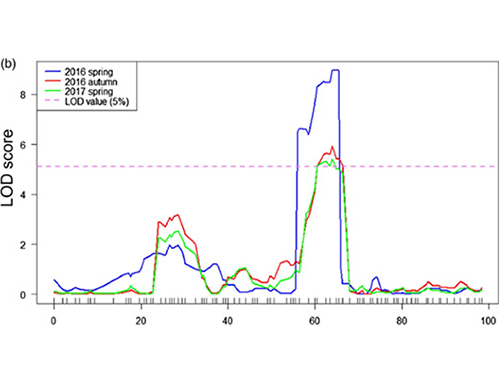 LOD mbiri dzegaka-huro-kureba QTL yakaonekwa paChr07 |
Xu, X., nevamwe."Iyo huru-effect quantitative trait locus Fnl7.1 inovhara kunonoka embryogenesis yakawanda mapuroteni ane hukama nehurefu hwehuro mucucumber."Plant Biotechnology Journal 18.7(2020).













