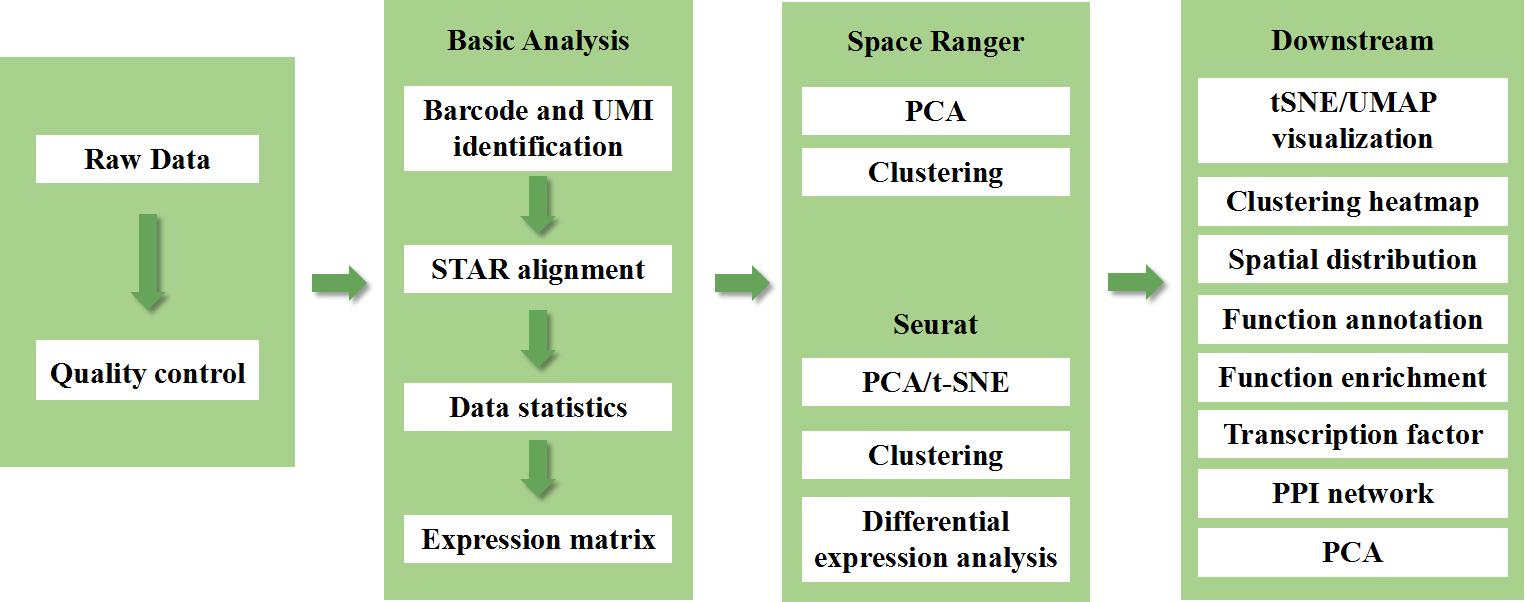
BMKMANU S1000 Spatial Transcriptome
BMKMANU S1000 Spatial Transcriptome Technical Scheme
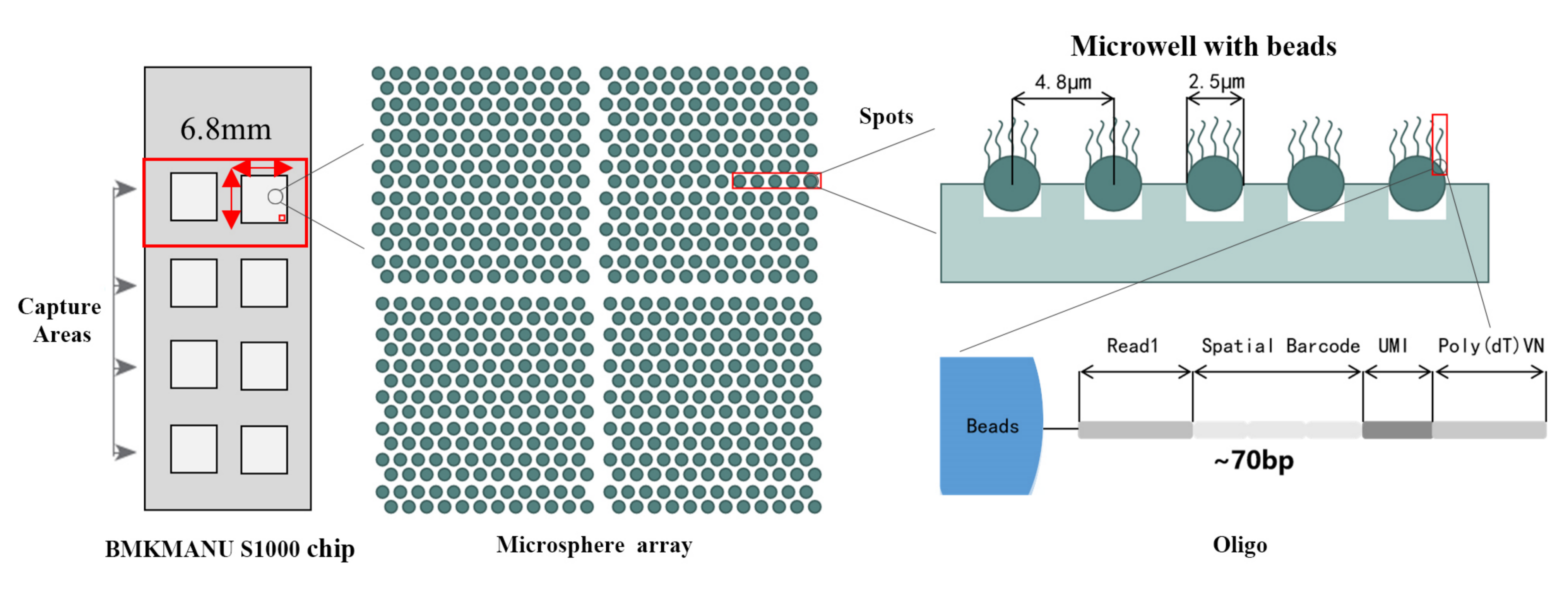
Zvakanakira zveBMKMANU S1000
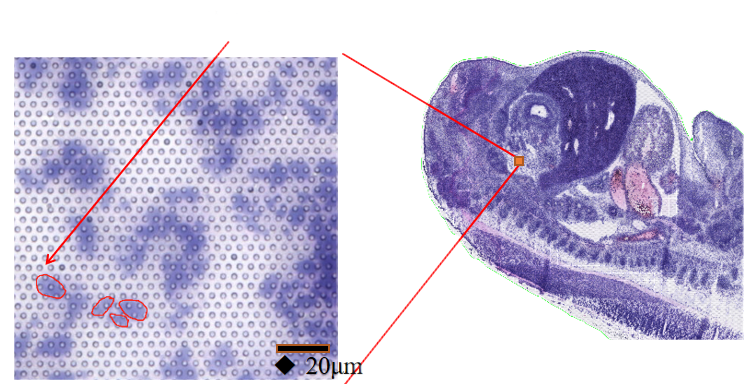
1) Sub-cellular resolution: Nzvimbo yega yega yekutora yaive > 2 miriyoni spatial Barcoded Spots ine dhayamita ye2.5 µm uye nzvimbo ye5 µm pakati penzvimbo dzemavara, ichigonesa spatial transcriptome kuongororwa ne sub-cellular resolution (5 µm).
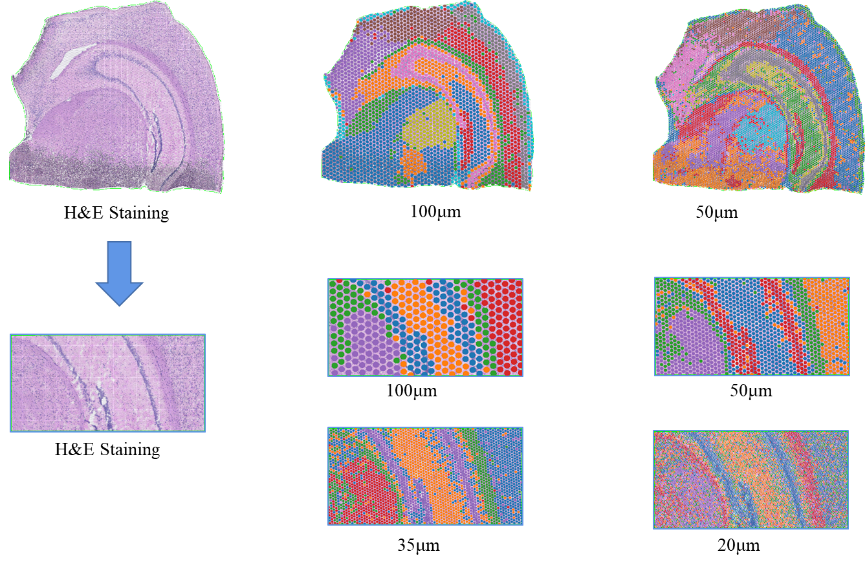
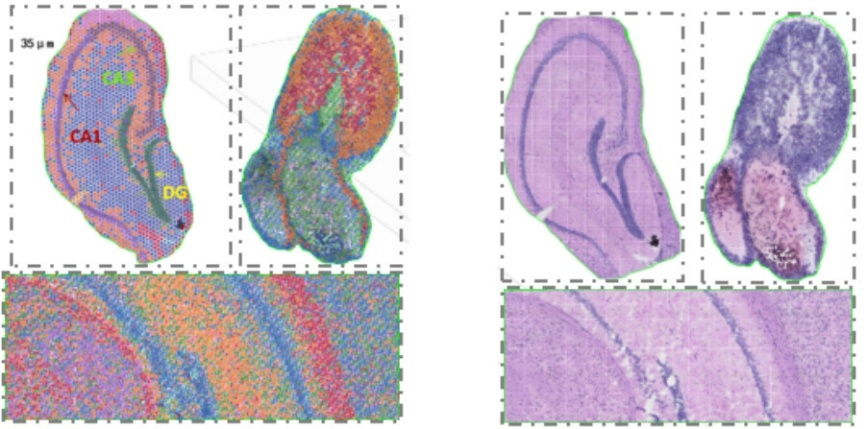
2) Multi-level resolution analysis: Flexible multi-level analysis kubva ku100 μm kusvika ku5 μm kugadzirisa zvakasiyana-siyana zvenyama pakugadzirisa kwakakwana.
3) Comprehensive transcriptome profiling: Zvinyorwa zvakatorwa kubva kune yese tissue slide zvinogona kuongororwa, pasina kudziviswa pahuwandu hwechinangwa chemajini uye nzvimbo inotarirwa.
Sevhisi Zvinotsanangurwa
| Library | Sequencing strategy | Data Output inokurudzirwa |
| S1000 cDNA raibhurari | BMKMANU S1000-Illumina PE150 | 60Gb / muenzaniso |
Sample Zvinodiwa
| Muenzaniso | Number | Size | RNA Hunhu |
| OCT yakamisikidzwa tishu block | 2-3 zvidhinha / muenzaniso | Approx.6.8x6.8x6.8 mm3 | RIN≥7 |
Kuti uwane rumwe ruzivo nezve nhungamiro yegadziriro yemuenzaniso uye mafambiro ebasa, ndapota inzwa wakasununguka kutaura kune aBMKGENE nyanzvi
Kuyerera Kwebasa Rebasa

Kuedza kugadzira

Muenzaniso wekutumira

Pre-kuedza
Spatial barcoding

Kuvakwa kweraibhurari

Sequencing

Data analysis

Mushure mekutengesa masevhisi
Iyo data yakagadzirwa neBMKMANU S1000 inoongororwa uchishandisa software "BSTMatrix", iyo yakazvimiririra yakagadzirwa neBMKGENE, kusanganisira:
1) Gene kutaura matrix chizvarwa
2) HE mufananidzo kugadzirisa
3) Inoenderana neyakadzika yechitatu-bato software yekuongorora
4) Pamhepo "BSTViewer" inobatsira kuwana mhedzisiro yekuona pane zvakasiyana zvigadziriso.
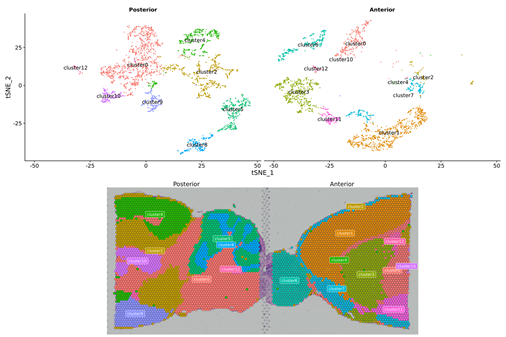
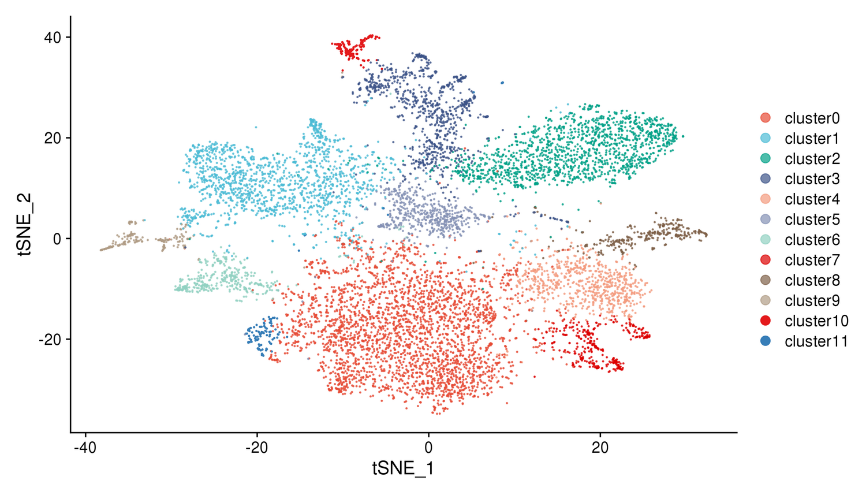
1.Spot clustering
2.Spatial distribution
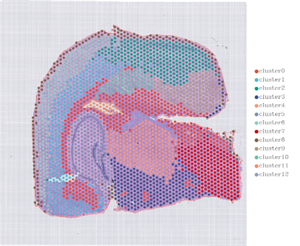
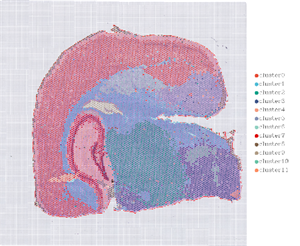
Note: Chisarudzolevel=13 (100 µm, ruboshwe); 7 (50 µm, rudyi)
3.Marker kutaura kuwanda kuunganidza heatmap
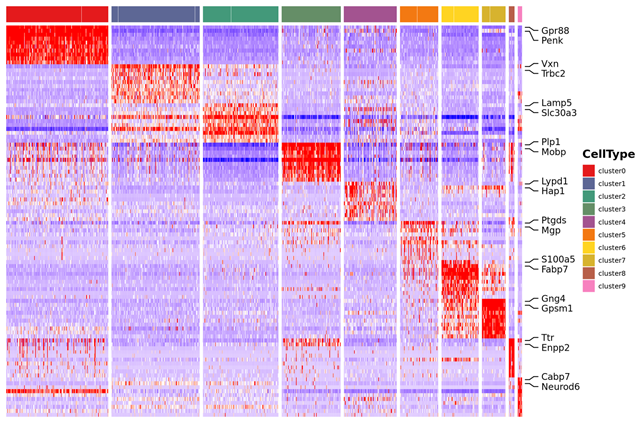
4.Inter-sample data analysis