Single- nucleus RNA Sequencing
Technical Scheme
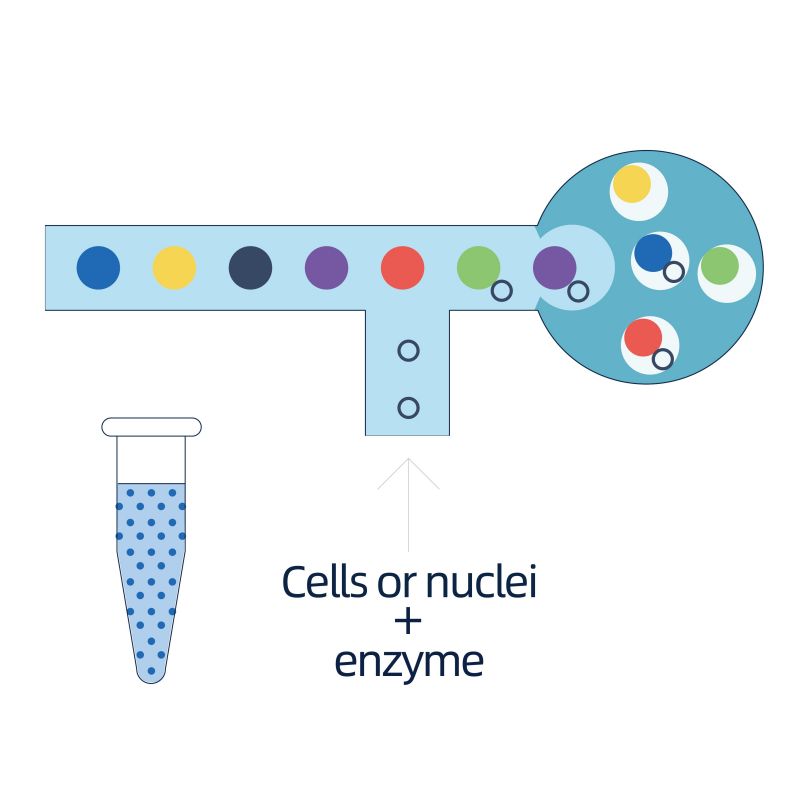
The isolation of nuclei is achieved by 10× Genomics ChromiumTM, which consists eight-channel microfluidics system with double crossings. In this system, a gel beads with barcodes and primer, enzymes and a single nucleus are encapsulated in nanoliter-sized oil drop, generating Gel Bead-in-Emulsion (GEM). Once GEM are formed, cell lysis and release of barcodes are performed in each GEM. mRNA are reverse transcribed into cDNA molecules with 10× barcodes and UMI, which are further subject to standard sequencing library construction.


Features
● Preparation of single-nuclei suspension from frozen tissues
● Formation of Gel Bead-in-Emulsion (GEM) followed by cDNA synthesis
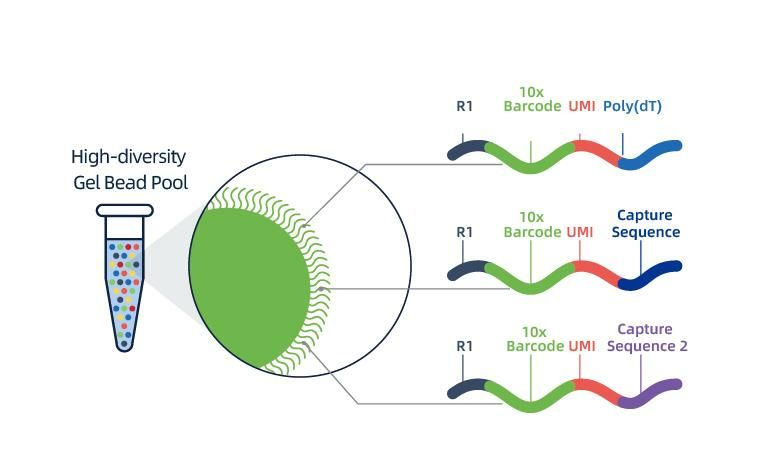
● Each bead in a GEM is loaded with primers composed of 4 sections:
poly(dT) tail for mRNA priming and cDNA synthesis,
Unique Molecular Identifier (UMI) to correct amplification bias
10x barcode
Binding sequence of partial read 1 sequencing primer
Advantages
Single-nucleus RNA sequencing circumvents the limitations of single-cell RNA sequencing, enabling:
● The use of frozen samples and not only limited to fresh samples
● Low stress of frozen cells when compared to enzymatic treatment of fresh cells, reflected in the transcriptome data in the form of less stress-induced genes
● No need for prior removal of red blood cells
● Unlimited cell diameter
● Large array of samples that are eligible for analysis, including complex and fragile tissue types that are prone to cell clumping or destruction during tissue dissociation
Samples that cannot be analysed by single-cell RNA sequencing and are eligible for single nuclei RNA sequencing:
|
Cell / Tissue |
Reason |
|
Unfresh frozen tissue |
Unable to get fresh or long-saved organizations |
|
Muscle cell, Megakaryocyte, Fat… |
Cell diameter is too large to enter the instrument |
|
Liver… |
Too fragile to break, unable to distinguish single cells |
|
Neuron cell, Brain… |
More sensitive, easy to stress, will change the sequencing results |
|
Pancreas, Thyroid… |
Rich in endogenous enzymes, affecting the production of single cell suspension |
Single-nucleus vs Single-cell
|
Single-nucleus |
Single-cell |
|
Unlimited cell diameter |
Cell diameter: 10-40 μm |
|
The material can be frozen tissue |
The material must be fresh tissue |
|
Low stress of frozen cells |
Enzyme treatment may cause cell stress reaction |
|
No red blood cells need to be removed |
Red blood cells need to be removed |
|
Nuclear expresses bioinformation |
The whole cell expresses bioinformation |
Specifications
|
Sample Requirements |
Library |
Sequencing strategy |
Data recommended |
Quality Control |
|
Cell number: total > 10^6 or > 5x105 flow sorted cells. Tissue > 200 mg Whole blood > 4 mL |
10x Genomics sn cDNA library |
Illumina PE150 |
100K PE reads per cell (100-200 Gb) |
700-1200 nuclei/μl and nuclei integrity observed under microscope |
For more details on sample preparation guidance and service workflow, please feel free to talk to a BMKGENE expert
Service Work Flow

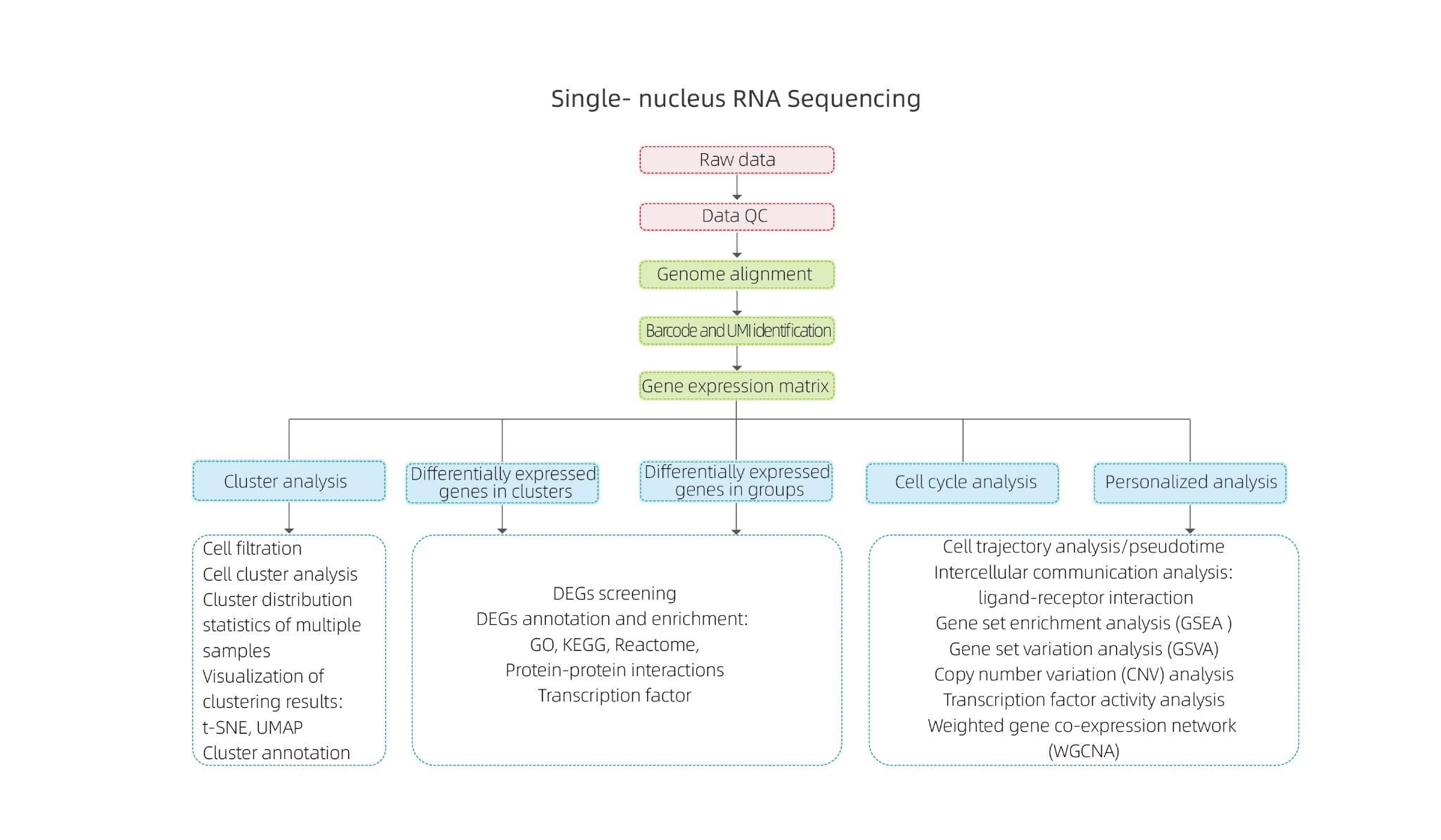
Includes the following analysis:
● Quality Control: number of cells, gene detection, accurate identification of cells, RNA molecules and expression quantification
● Inner Sample Analysis:
Cell clustering and cluster annotation
Differential Expression Analysis: identification of DEGs in clusters
Functional annotation and enrichment of cluster DEGs
● Inter-group Analysis:
Combination of data
Differential Expression Analysis: identification of DEGs in groups
Functional annotation and enrichment of group DEGs
● Advanced Analysis:
Cell cycle analysis
Pseudotime analysis
Cell communication analysis (CellPhoneDB)
Gene Set Enrichment Analysis (GSEA)
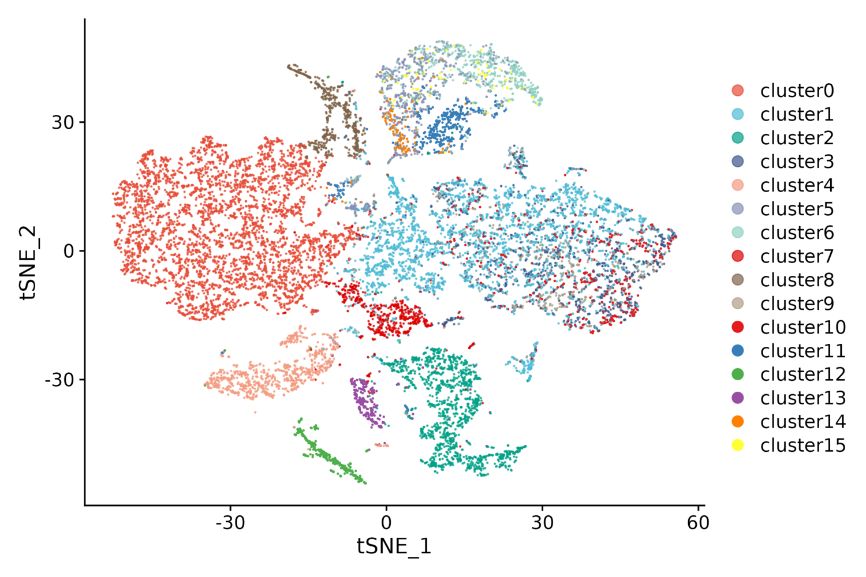
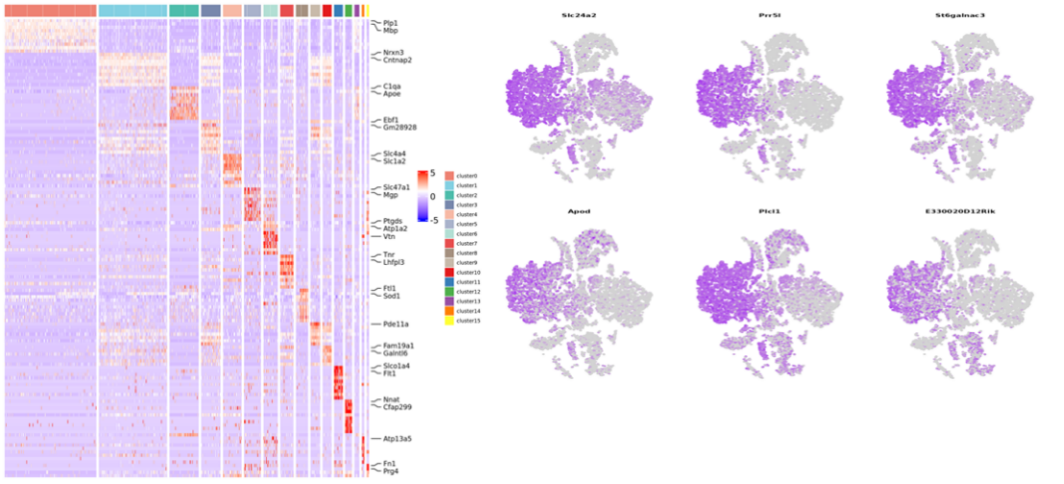
Inner-sample analysis
Cell clustering:
Differential Expression Analysis: cluster DEGs
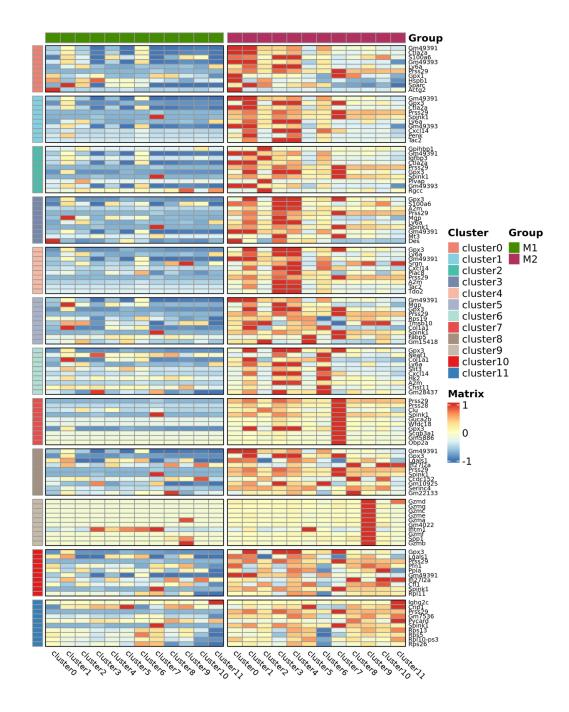
Inter-group analysis
Differential Expression Analysis: Group DEGs
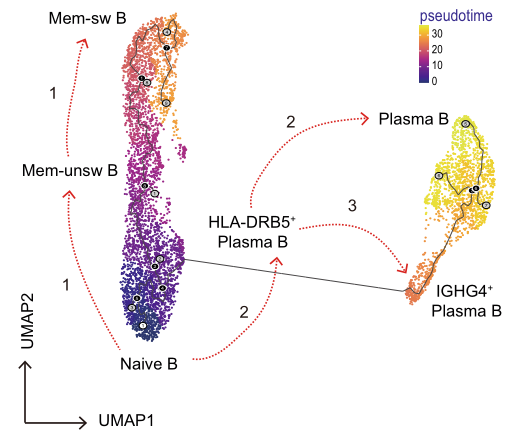
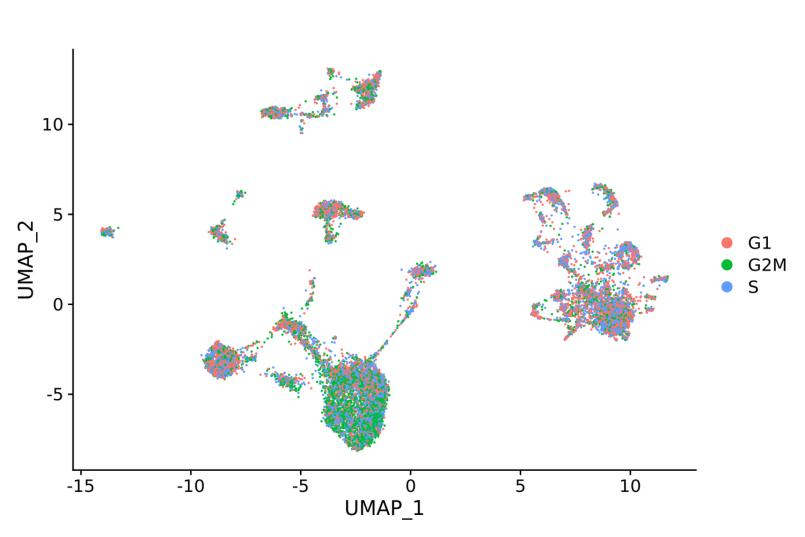
Advanced Analysis:
Pseudotime analysis:
Cell cycle analysis:
Explore the advancements facilitated by BMKGene’s single-nucleus RNA sequencing services by 10X Chromium in these featured publications:
Wang, L. et al. (2021) ‘Single-cell transcriptomic analysis reveals the immune landscape of lung in steroid-resistant asthma exacerbation’, Proceedings of the National Academy of Sciences of the United States of America, 118(2), p. e2005590118. doi: 10.1073/pnas.2005590118
Zheng, H. et al. (2022) ‘A Global Regulatory Network for Dysregulated Gene Expression and Abnormal Metabolic Signaling in Immune Cells in the Microenvironment of Graves’ Disease and Hashimoto’s Thyroiditis’, Frontiers in Immunology, 13, p. 879824. doi: 10.3389/FIMMU.2022.879824/BIBTEX.
Tian, H. et al. (2023) ‘Single-cell transcriptome uncovers heterogeneity and immune responses of leukocytes after vaccination with inactivated Edwardsiella tarda in flounder (Paralichthys olivaceus)’, Aquaculture, 566, p. 739238. doi: 10.1016/J.AQUACULTURE.2023.739238.
Yu, Y. et al. (2023) ‘Photodynamic therapy improves the outcome of immune checkpoint inhibitors via remodelling anti-tumour immunity in patients with gastric cancer’, Gastric Cancer, 26(5), pp. 798–813. doi: 10.1007/S10120-023-01409-X/METRICS.