Kutsata kwanthawi yayitali kosalemba-Illumina
Ubwino wa Utumiki
● Ubwino wa Utumiki
● Ma cell ndi minofu yeniyeni
● Gawo lachindunji likuwonekera ndikuwonetsa kusintha kwachiwonetsero
● Njira yeniyeni ya nthawi ndi malo
● Kusanthula kophatikizana ndi data ya mRNA.
● BMKCloud yochokera ku zotsatira zobweretsera: Customize data-mining kupezeka pa nsanja.
● Ntchito zogulitsa pambuyo pogulitsa ndizovomerezeka kwa miyezi itatu polojekiti ikamalizidwa
Zitsanzo Zofunika ndi Kutumiza
| Library | nsanja | Deta yovomerezeka | Zithunzi za QC |
| Kusintha kwa rRNA | Chithunzi cha PE150 | 10 gb | Q30≥85% |
| Conc.(ng/μl) | Mtengo (μg) | Chiyero | Umphumphu |
| ≥100 | ≥ 0.5 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Zochepa kapena zopanda mapuloteni kapena DNA kuipitsidwa komwe kumawonetsedwa pa gel. | Kwa zomera: RIN≥6.5; Kwa nyama: RIN≥7.0; 5.0≥28S/18S≥1.0; kukwera kochepa kapena kopanda koyambira |
Nucleotides:
Thupi: Kulemera kwake (kuuma): ≥1 g
*Pa minofu yaing'ono kuposa 5 mg, timalimbikitsa kutumiza zitsanzo za minofu yowuma (mu madzi a nayitrogeni).
Kuyimitsidwa kwa ma cell: kuchuluka kwa ma cell = 3 × 107
*Tikupangira kutumiza ma cell a frozen lysate.Ngati seloyo imawerengera yaying'ono kuposa 5 × 105, kung'anima kozizira mumadzi a nayitrogeni akulimbikitsidwa.
Zitsanzo za magazi:
PA×geneBloodRNATube;
6mLTRIzol ndi 2mL magazi(TRIzol:Blood=3:1)
Kutumiza Kwachitsanzo Kovomerezeka
Chidebe: 2 ml centrifuge chubu (zojambula za malata ndizosavomerezeka)
Zitsanzo zolembera: Gulu+ lofanana mwachitsanzo A1, A2, A3;B1, B2, B3...
Kutumiza:
1.Dry-ice: Zitsanzo ziyenera kuikidwa m'matumba ndikukwiriridwa mu ayezi wouma.
2.RNAstable machubu: RNA zitsanzo akhoza zouma mu RNA kukhazikika chubu (mwachitsanzo RNAstable®) ndi kutumizidwa mu firiji.
Kuyenda kwa Ntchito Yantchito

Kuyesera kupanga

Kupereka zitsanzo

Kusintha kwa RNA

Kumanga laibulale

Kutsata

Kusanthula deta

Pambuyo pogulitsa ntchito
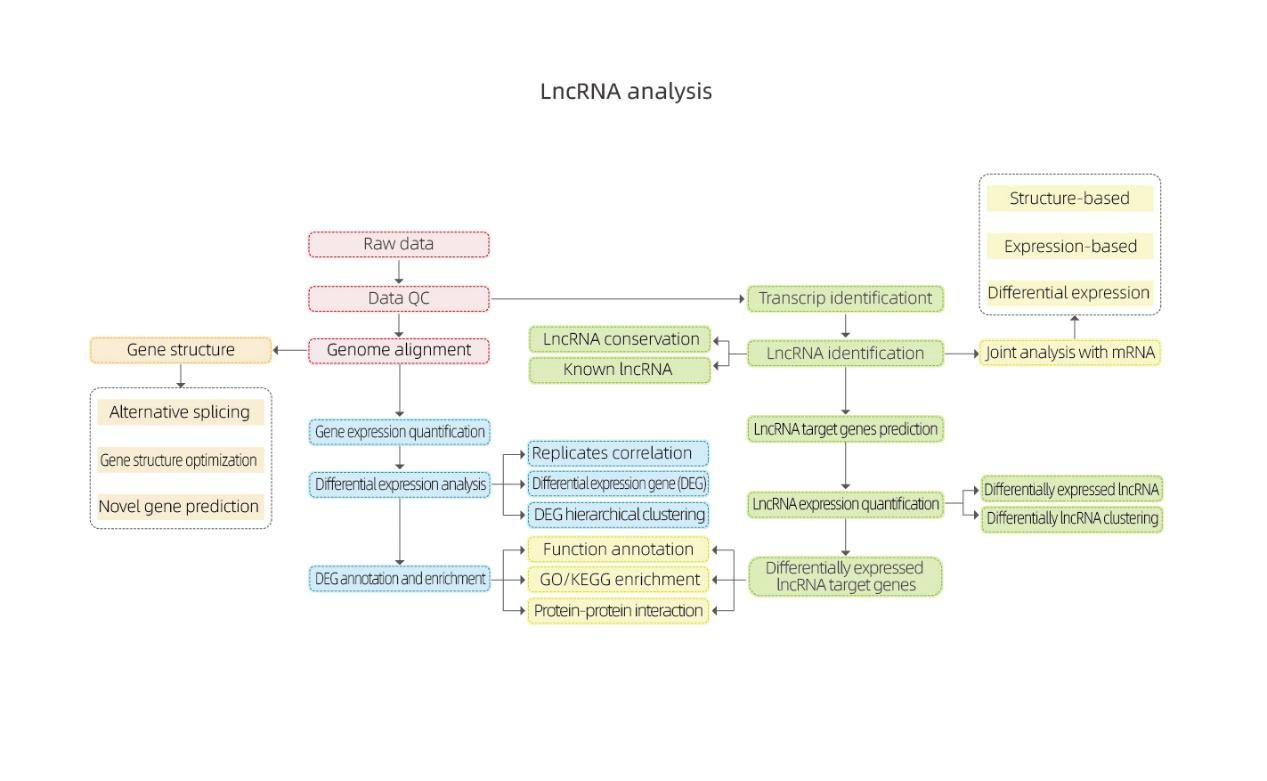
Bioinformatics
Gulu la 1.LncRNA
LncRNA yonenedweratu ndi mapulogalamu anayi omwe ali pamwambawa adagawidwa m'magulu a 4: lincRNA, anti-sense-LncRNA, intronic-LncRNA;malingaliro-LncRNA.Gulu la LncRNA likuwonetsedwa mu histogram pansipa.
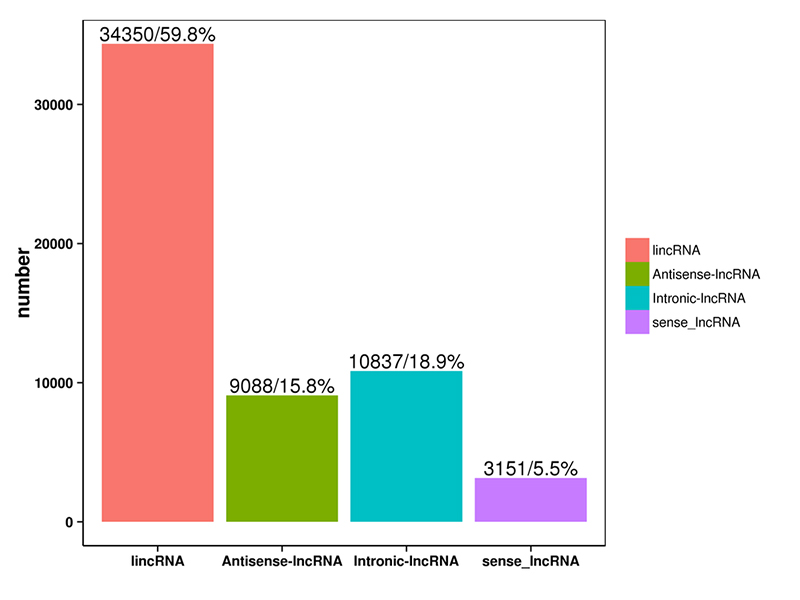
Gawo la LncRNA
2.Cis-targeted genes of DE-lncRNA enrichment analysis
ClusterProfiler anagwiritsidwa ntchito mu GO enrichment analysis pa cis-targeted gene of differentially lncRNA (DE-lncRNA), ponena za biological process, molecular function and cellular components.Kusanthula kwa GO enrichment ndi njira yodziwira mawu opangidwa ndi DEG olemeretsedwa kwambiri poyerekeza ndi ma genome onse.Mawu olemeretsedwa anaperekedwa mu histogram, bubble chart, etc. monga momwe zilili pansipa.
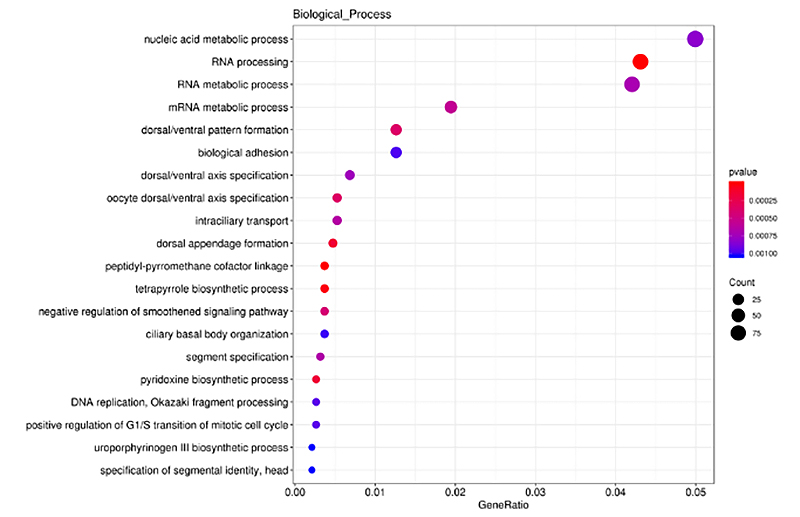 Cis-targeted genes of DE-lncRNA enrichment analysis -Bubble chart
Cis-targeted genes of DE-lncRNA enrichment analysis -Bubble chart
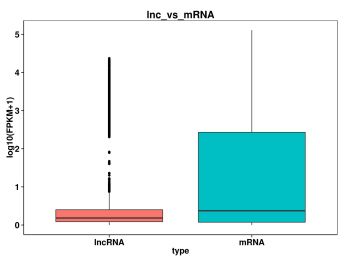
3. Poyerekeza kutalika, nambala ya exon, ORF ndi kuchuluka kwa mawu a mRNA ndi lncRNA, tikhoza kumvetsetsa kusiyana kwa mapangidwe, ndondomeko ndi zina zotero pakati pawo, ndikutsimikiziranso ngati buku la lncRNA lomwe linanenedweratu ndi ife likugwirizana ndi makhalidwe onse.
BMK Case
Mbiri ya lncRNA yochepetsedwa mu mbewa mapapo adenocarcinomas yokhala ndi kusintha kwa KRAS-G12D ndi kugogoda kwa P53
Lofalitsidwa:Journal of Cellular ndi Molecular Medicine,2019
Njira yotsatirira
Ilumina
Zosonkhanitsa zitsanzo
Maselo a NONMMUT015812-knockdown KP (shRNA-2) ndi ma cell control (sh-Scr) omwe adapezeka pa tsiku la 6 la matenda enaake a virus.
Zotsatira zazikulu
Kafukufukuyu amafufuza ma lncRNA owonetsedwa molakwika mu mbewa lung adenocarcinoma yokhala ndi kugogoda kwa P53 ndi kusintha kwa KrasG12D.
1.6424 lncRNAs adawonetsedwa mosiyana (≥ 2-fold kusintha, P <0.05).
2.Pakati pa 210 lncRNAs (FC≥8), mawu a 11 lncRNAs adayendetsedwa ndi P53, 33 lncRNAs ndi KRAS ndi 13 lncRNAs ndi hypoxia m'maselo oyambirira a KP, motero.
3.NONMMUT015812, yomwe inakhazikitsidwa modabwitsa mu mouse lung adenocarcinoma ndipo inayendetsedwa molakwika ndi P53 re-expression, inadziwika kuti ifufuze ntchito yake yama cell.
4.Kugwetsa kwa NONMMUT015812 ndi shRNAs kunachepetsa kuchulukira ndi kusamuka kwa maselo a KP.NONMMUT015812 anali oncogene wotheka.
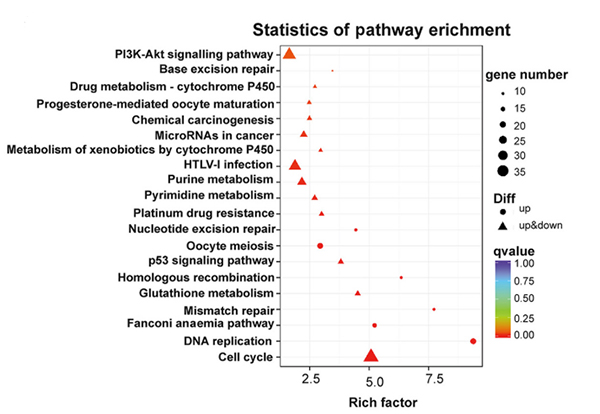 KEGG njira yowunikira ma jini owonetsedwa mosiyanasiyana m'maselo a NONMMUT015812-knockdown KP | 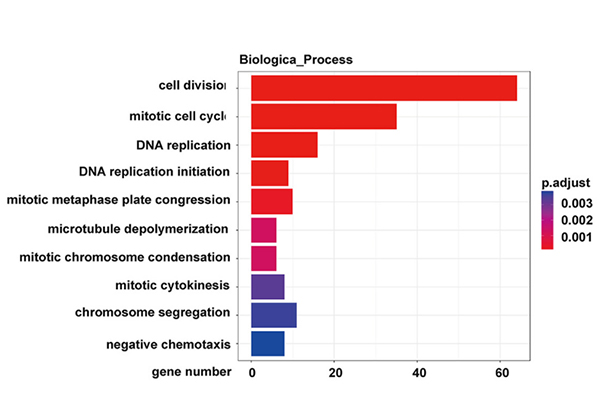 Kusanthula kwa Gene Ontology kwa majini owonetsedwa mosiyanasiyana m'maselo a NONMMUT015812-knockdown KP |
Buku
Mbiri ya lncRNA yochepetsedwa mu mbewa mapapo adenocarcinomas yokhala ndi kusintha kwa KRAS-G12D ndi kugogoda kwa P53 [J].Journal of Cellular and Molecular Medicine, 2019, 23(10).DOI: 10.1111/jcmm.14584