Metatranscriptome Sequencing
Service Features
● rRNA depletion followed by directional mRNA library preparation.
● Sequencing on Illumina NovaSeq.
Service Advantages
● Study the Changes of Complex Microbial Communities: This happens at the transcriptional level and explore potential new genes.
● Explaining Microbial Community Interactions with the Host or Environment.
● Comprehensive Bioinformatic Analysis: This provides insights on community taxonomic and functional compositions, as well as differential gene expression analysis.
● Extensive Gene Annotation: Using up-to-date gene function databases for informative gene expression information of microbial communities.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Service Specifications
|
Sequencing platform |
Sequencing Strategy |
Data recommended |
Data Quality Control |
|
Illumina NovaSeq |
PE150 |
12Gb |
Q30≥85% |
Sample Requirements
|
Concentration (ng/µL) |
Total amount (µg) |
Volume (µL) |
OD260/280 |
OD260/230 |
RIN |
|
≥50 |
≥0.5 |
≥20 |
1.6-2.5 |
1.0-2.5 |
≥6.5 |
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
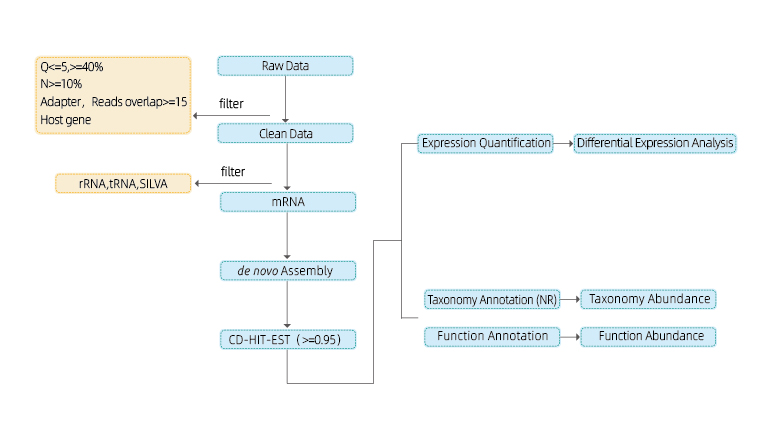
Includes the following analysis:
● Sequencing Data Quality Control
● Transcript Assembly
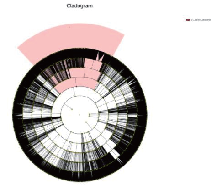
● Taxonomic Annotation and Abundance
● Functional Annotation and Abundance
● Expression Quantification and Differential Analysis
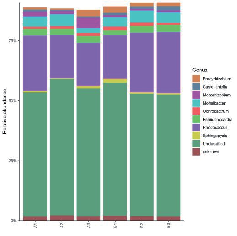
Taxonomic distribution of each sample:
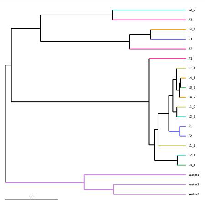
Beta diversity analysis: UPGMA
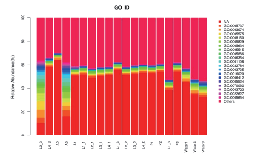
Functional annotation – GO abundance
Differential taxonomy abundance – LEFSE
Explore the advancements facilitated by BMKGene’s meta transcriptomics sequencing services through a curated collection of publications.
Lu, Z. et al. (2023) ‘Acid tolerance of lactate-utilizing bacteria of the order Bacteroidales contributes to prevention of ruminal acidosis in goats adapted to a high-concentrate diet’, Animal Nutrition, 14, pp. 130–140. doi: 10.1016/J.ANINU.2023.05.006.
Song, Z. et al. (2017) ‘Unraveling core functional microbiota in traditional solid-state fermentation by high-throughput amplicons and metatranscriptomics sequencing’, Frontiers in Microbiology, 8(JUL). doi: 10.3389/FMICB.2017.01294/FULL.
Wang, W. et al. (2022) ‘Novel Mycoviruses Discovered from a Metatranscriptomics Survey of the Phytopathogenic Alternaria Fungus’, Viruses, 14(11), p. 2552. doi: 10.3390/V14112552/S1.
Wei, J. et al. (2022) ‘Parallel metatranscriptome analysis reveals degradation of plant secondary metabolites by beetles and their gut symbionts’, Molecular Ecology, 31(15), pp. 3999–4016. doi: 10.1111/MEC.16557.