Long Non-coding Sequencing-Illumina
Service Advantages
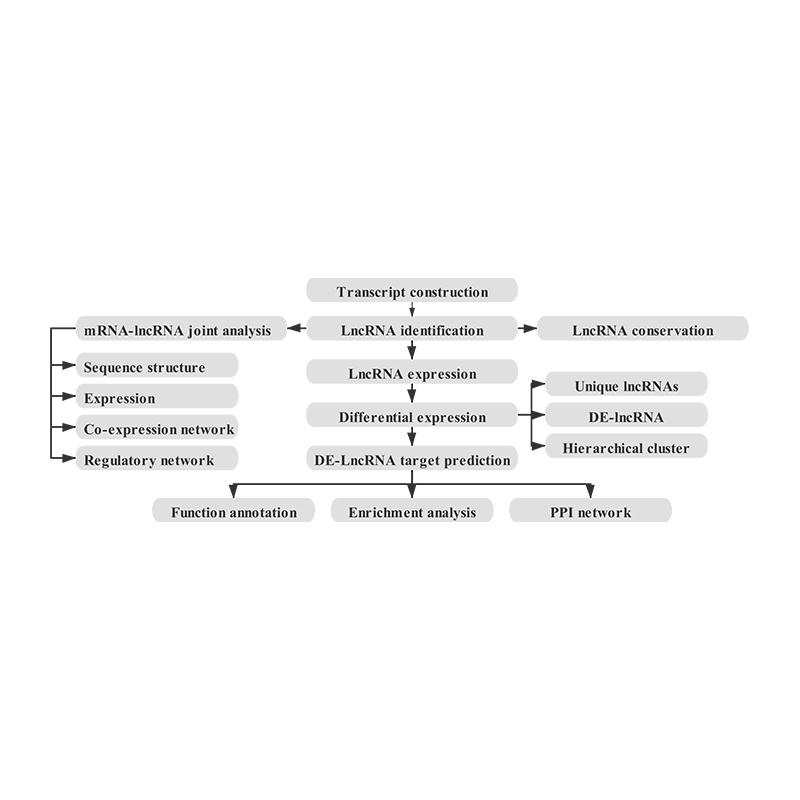
● Joint analysis of mRNA and lncRNA: by combining the quantification of mRNA transcripts with the study of lncRNA and their targets, it is possible to get an in-depth overview of the regulatory mechanism underlying the cellular response.
● Extensive Expertise: Our team brings a wealth of experience to every project, with a track record of processing over 20,000 samples at BMK spanning diverse sample types and lncRNA projects.
● Rigorous Quality Control: We implement core control points across all stages, from sample and library preparation to sequencing and bioinformatics. This meticulous monitoring ensures the delivery of consistently high-quality results.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Sample Requirements and Delivery
|
Library |
Platform |
Recommended data |
Data QC |
|
rRNA depleted directional library |
Illumina PE150 |
10-16 Gb |
Q30≥85% |
Nucleotides:
|
Conc.(ng/μl) |
Amount (μg) |
Purity |
Integrity |
|
≥ 80 |
≥ 0.5 |
OD260/280=1.7-2.5 OD260/230=0.5-2.5 Limited or no protein or DNA contamination shown on gel. |
For plants: RIN≥6.5; For animals: RIN≥7.0; 5.0≥28S/18S≥1.0; limited or no baseline elevation |
● Plants:
Root, Stem or Petal: 450 mg
Leaf or Seed: 300 mg
Fruit: 1.2 g
● Animal:
Heart or Intestine: 450 mg
Viscera or Brain: 240 mg
Muscle: 600 mg
Bones, Hair or Skin: 1.5g
● Arthropods:
Insects: 9g
Crustacea: 450 mg
● Whole blood: 2 tubes
● Cells: 106 cells
● Serum and Plasma: 6 mL
Recommended Sample Delivery
Container: 2 ml centrifuge tube (Tin foil is not recommended)
Sample labeling: Group+replicate e.g. A1, A2, A3; B1, B2, B3.
Shipment:
1. Dry-ice: Samples need to be packed in bags and buried in dry-ice.
2. RNAstable tubes: RNA samples can be dried in RNA stabilization tube(e.g. RNAstable®) and shipped in room temperature.
Service Work Flow

Experiment design

Sample delivery

RNA extraction

Library construction

Sequencing

Data analysis

After-sale services
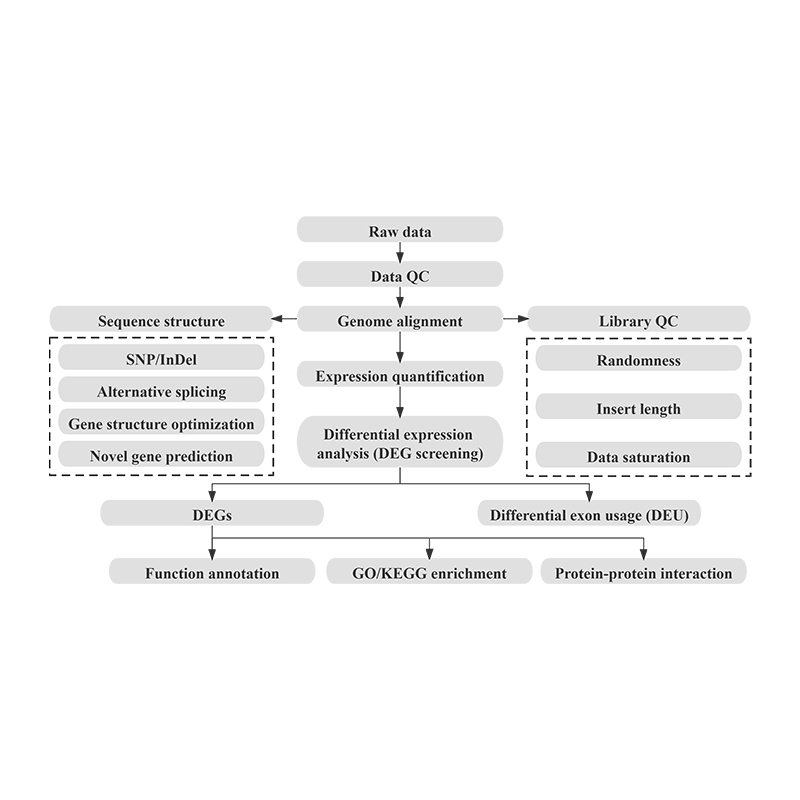
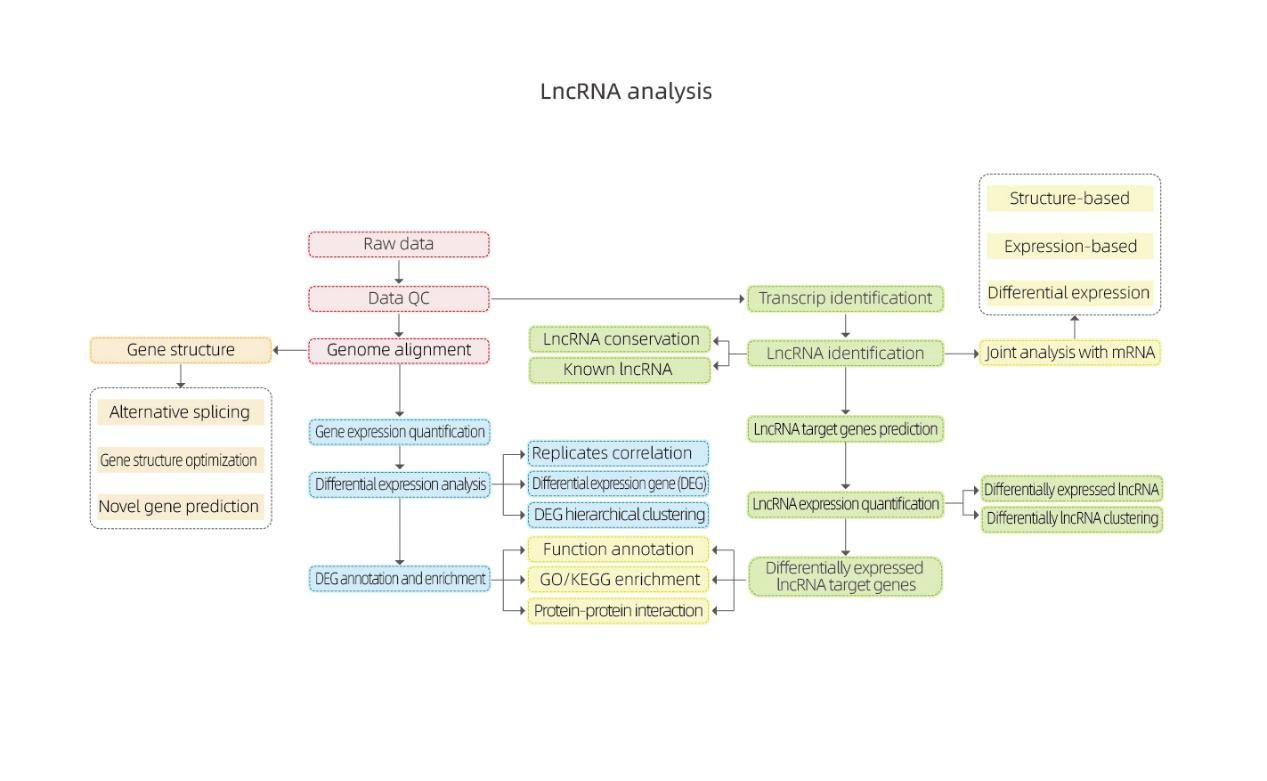
Bioinformatics
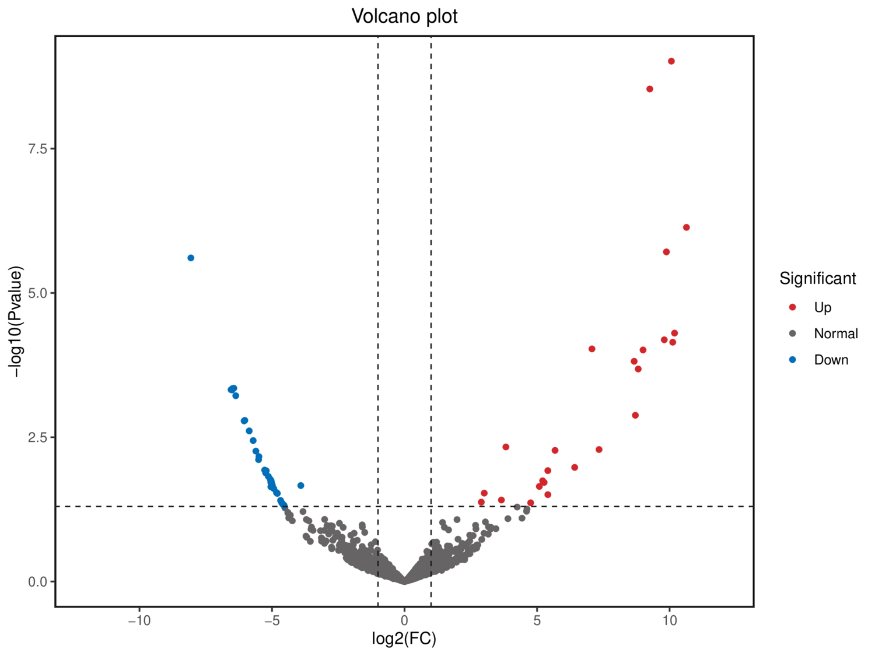
Differential Gene Expression (DEGs) analysis
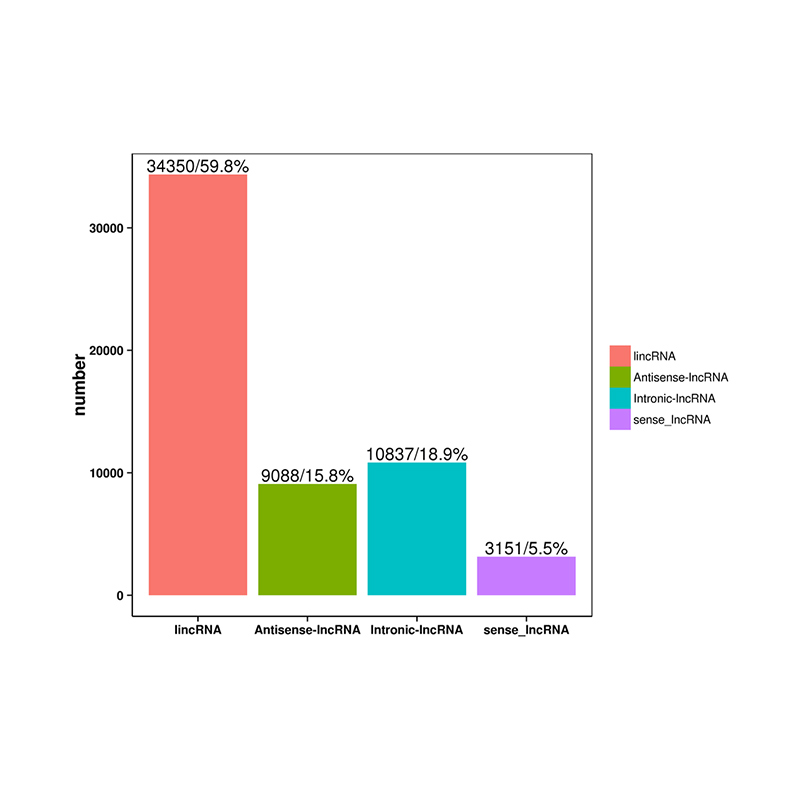
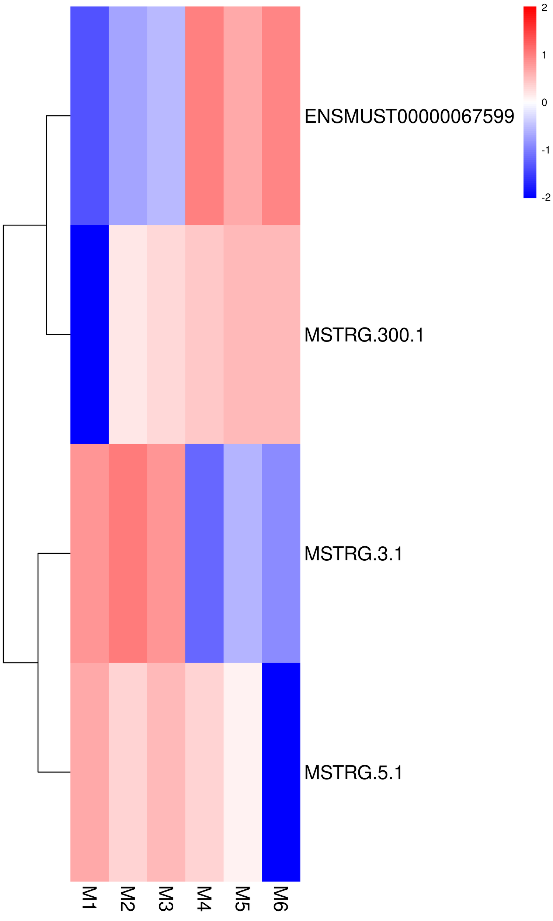
Quantification of lncRNA expression – clustering
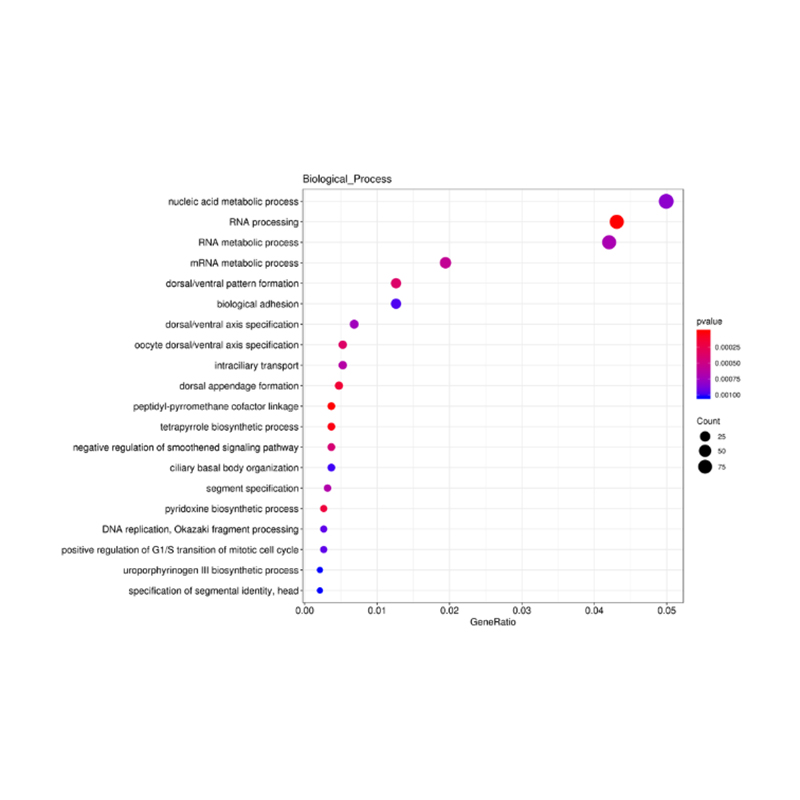
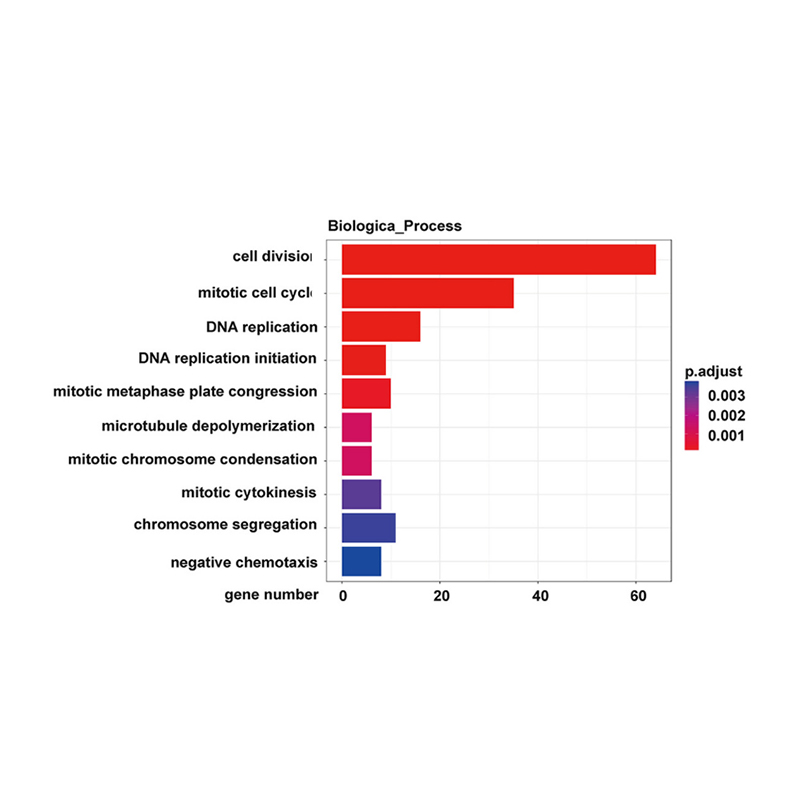
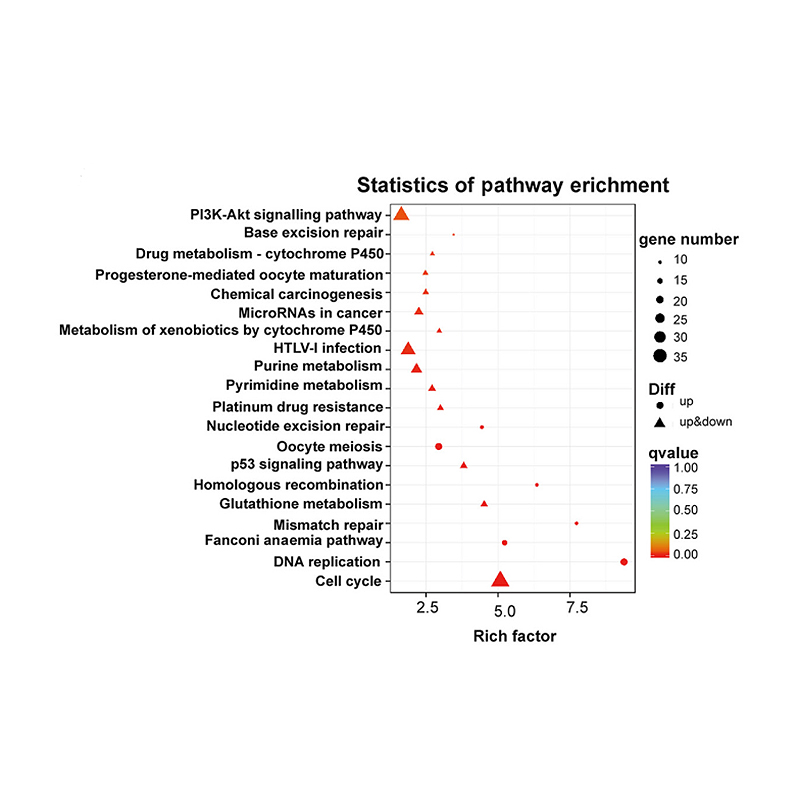
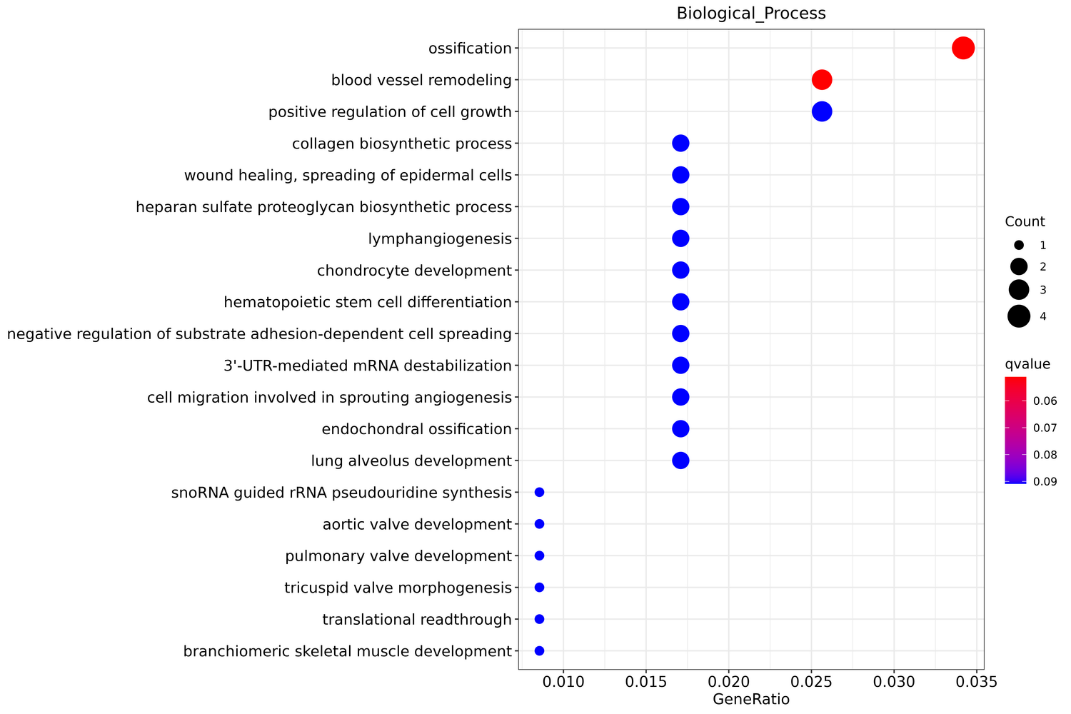
Enrichment of lncRNA target genes
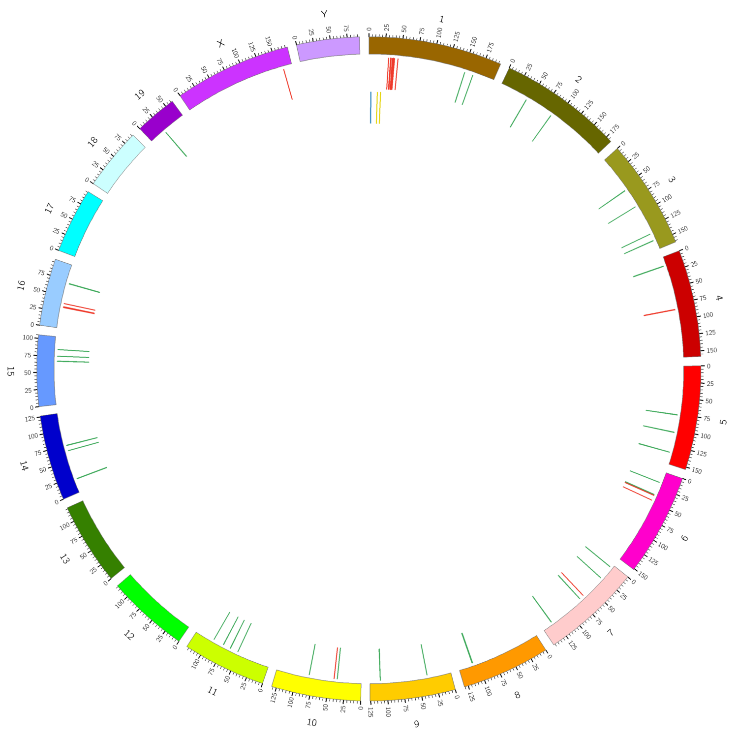
Joint mRNA and lncRNA position analysis – Circos plot (middle circle is mRNA and inner cicrlce is lncRNA)
Explore the advancements facilitated by BMKGene’s lncRNA equencing services through a curated collection of publications.
Ji, H. et al. (2020) ‘Identification, functional prediction, and key lncRNA verification of cold stress-related lncRNAs in rats liver’, Scientific Reports 2020 10:1, 10(1), pp. 1–14. doi: 10.1038/s41598-020-57451-7.
Jia, Z. et al. (2021) ‘Integrative Transcriptomic Analysis Reveals the Immune Mechanism for a CyHV-3-Resistant Common Carp Strain’, Frontiers in Immunology, 12, p. 687151. doi: 10.3389/FIMMU.2021.687151/BIBTEX.
Wang, X. J. et al. (2022) ‘Multi-Omics Integration-Based Prioritisation of Competing Endogenous RNA Regulation Networks in Small Cell Lung Cancer: Molecular Characteristics and Drug Candidates’, Frontiers in Oncology, 12, p. 904865. doi: 10.3389/FONC.2022.904865/BIBTEX.
Xiao, L. et al. (2020) ‘Genetic dissection of the gene coexpression network underlying photosynthesis in Populus’, Plant Biotechnology Journal, 18(4), pp. 1015–1026. doi: 10.1111/PBI.13270.
Zheng, H. et al. (2022) ‘A Global Regulatory Network for Dysregulated Gene Expression and Abnormal Metabolic Signaling in Immune Cells in the Microenvironment of Graves’ Disease and Hashimoto’s Thyroiditis’, Frontiers in Immunology, 13, p. 879824. doi: 10.3389/FIMMU.2022.879824/BIBTEX.