T2T GENOME ASSEMBY, GAP FREE GENOME
1stMkpụrụ osisi Rice abụọ1
Isiokwu: Mgbakọ na nkwado nke mkpụrụ ndụ ihe nketa abụọ na-enweghị oghere maka Xian/indica Rice na-ekpughe nghọta n'ime ihe owuwu osisi Centromere
Doi:https://doi.org/10.1101/2020.12.24.424073
Oge ebiputere: Jenụwarị 01, 2021.
Ụlọ ọrụ: Huazhong Agricultural University, China
Akụrụngwa
O. sativa xian/indicaỤdị osikapa 'Zhenshan 97 (ZS97)' na 'Minghui 63 (MH63)
Usoro usoro
NGS na-agụ + HiFi na-agụ + CLR na-agụ + BioNano + Hi-C
Data:
ZS97: 8.34 Gb (~ 23x) HiFi na-agụ + 48.39 Gb (~ 131x) CLR na-agụ + 25 Gb (~ 69x) NGS + 2 sel BioNano Irys
MH63: 37.88 Gb (~ 103x) HiFi na-agụ + 48.97 Gb (~ 132x) CLR na-agụ + 28 Gb(~76x) NGS + 2 BioNano Irys cell
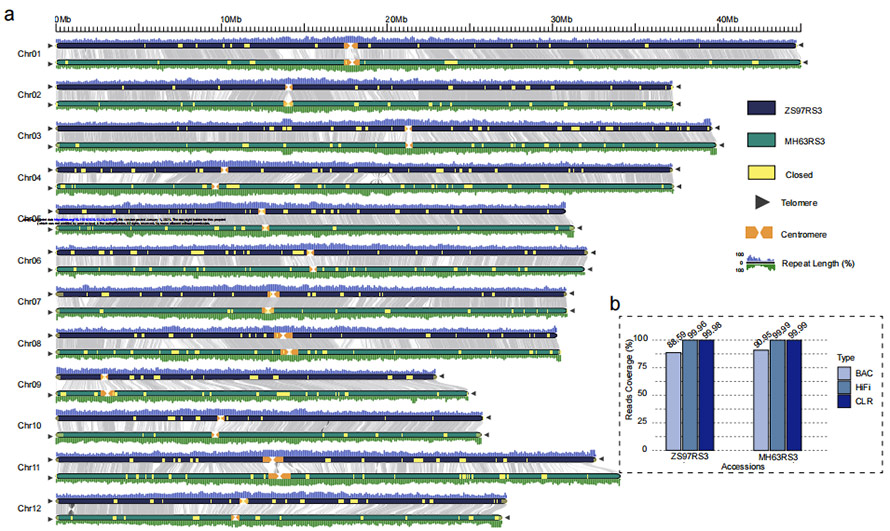
Ọgụgụ 1 Genomy osikapa abụọ na-enweghị oghere (MH63 na ZS97)
2ndBanana Genome2
Aha: Telomere-to-telomere chromosomes nke banana enweghị oghere na-eji usoro nanopore
Doi:https://doi.org/10.1101/2021.04.16.440017
Oge ebiputere: Eprel 17, 2021.
Ụlọ akwụkwọ: Mahadum Paris-Saclay, France
Akụrụngwa
Haploid okpukpu abụọMusa acuminatasppmalaccensis(DH-Pahang)
Atụmatụ usoro na data:
Ọnọdụ HiSeq2500 PE250 + MinION/ PromethION (93Gb, ~ 200X)+ Maapụ anya (DLE-1+BspQ1)
Isiokwu 1 Ntụnyere Musa acuminata (DH-Pahang) genome mgbakọ
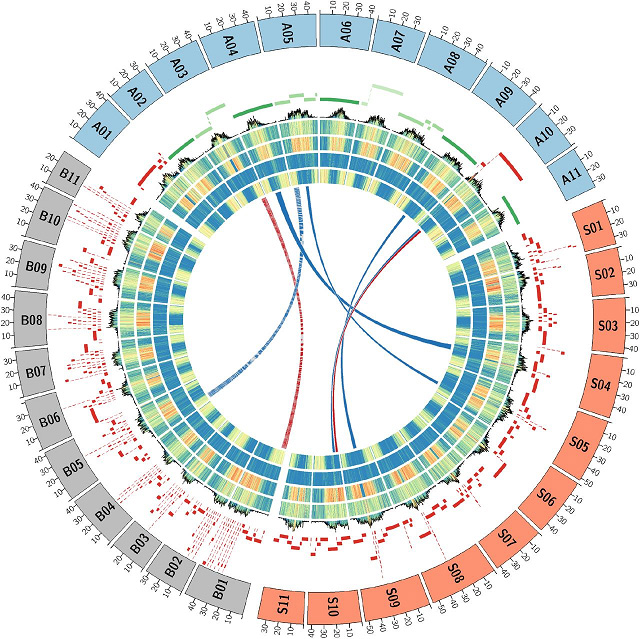
Ọgụgụ 2 Musa genomes architecture ntụnyere
3rdPheodactylum tricornutum genome3
Mbụ: Telomere-to-telomere genome Assembly ofP
haeodactylum tricornutum
Doi:https://doi.org/10.1101/2021.05.04.442596
Oge ebiputere: Mee 04, 2021
Ụlọ akwụkwọ: Western University, Canada
Akụrụngwa
Pheodactylum tricornutum(Nchịkọta Omenala nke Algae na Protozoa CCAP 1055/1)
Atụmatụ usoro na data:
1 Oxford Nanopore minION na-erugharị cell + 2 × 75 jikọtara ngwụcha ngwụcha ngwụcha Seq 550 na-agba ọsọ
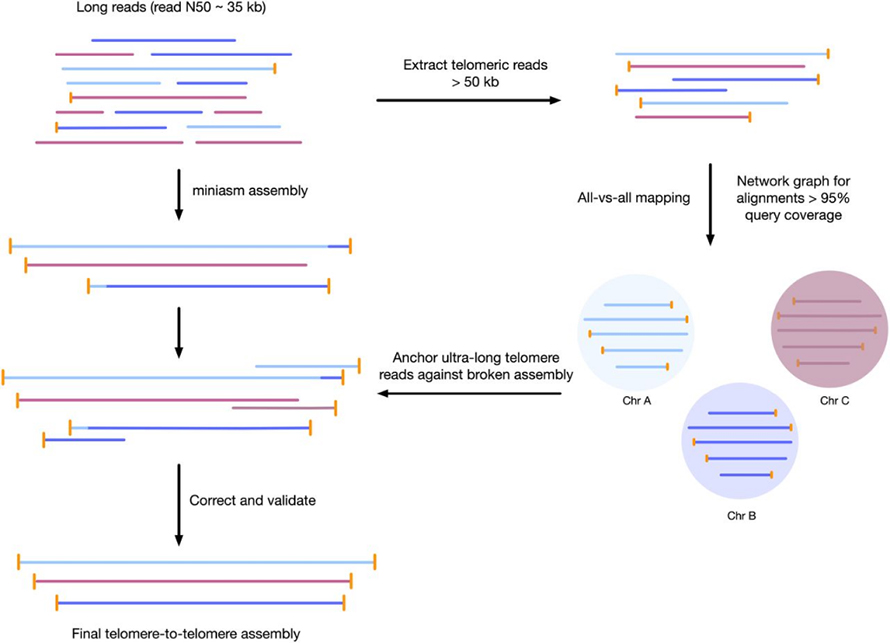
Ọgụgụ 3 Nrụ ọrụ maka mgbakọ telomere-to-telomere genome
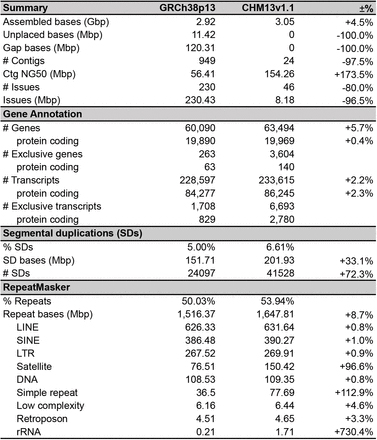
4thgenome nke mmadụ CHM134
Isiokwu: Usoro zuru oke nke genome mmadụ
Doi:https://doi.org/10.1101/2021.05.26.445798
Oge ebiputere: Mee 27, 2021
Ụlọ ọrụ: Ụlọ Ọrụ Na-ahụ Maka Ahụ Ike Mba (NIH), USA
Akụrụngwa: ahịrị cell CHM13
Atụmatụ usoro na data:
30 × PacBio okirikiri consensus sequencing (HiFi), 120 × Oxford Nanopore ultra-long read sequencing, 100 × Illumina PCR-Free sequencing (ILMN) , 70 × Illumina / Arima Genomics Hi-C (Hi-C), BioNano ngwa anya map, na Strand-seq
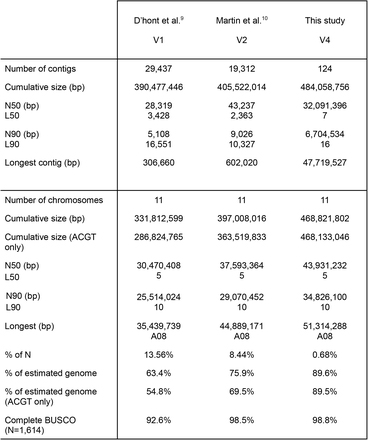
Tebụl 2 Ntụle nke GRCh38 na T2T-CHM13 mgbakọ genome mmadụ
Ntụaka
1.Sergey Nurk et al.Usoro zuru oke nke genome mmadụ.bioRxiv 2021.05.26.445798;doi:https://doi.org/10.1101/2021.05.26.445798
2. Caroline Belser et al.Telomere-to-telomere chromosomes enweghị oghere nke banana na-eji usoro nanopore.bioRxiv 2021.04.16.440017;doi:https://doi.org/10.1101/2021.04.16.440017
3.Daniel J. Giguere et al.Mgbakọ genome nke Telomere-to-telomere nke Phaeodactylum tricornutum.bioRxiv 2021.05.04.442596;doi:https://doi.org/10.1101/2021.05.04.442596
4.Jia-Ming Song et al.Mgbakọ na nkwado nke mkpụrụ ndụ ihe nketa na-enweghị oghere abụọ maka Xian/indica Rice na-ekpughere nghọta n'ime ihe owuwu osisi Centromere.bioRxiv 2020.12.24.424073;doi:https://doi.org/10.1101/2020.12.24.424073
Oge nzipu: Jan-06-2022

