T2T GENOME Asanble, GAP GRATIS GENOME
1stDe jenòm diri1
Tit: Asanble ak Validasyon de jenom referans san diferans pou diri Xian/indica revele apèsi sou Achitekti Centromere Plant
Doi:https://doi.org/10.1101/2020.12.24.424073
Lè afiche: 01 janvye 2021.
Enstiti: Huazhong Agricultural University, Lachin
Materyèl
O. sativa xian/indicavaryete diri 'Zhenshan 97 (ZS97)' ak 'Minghui 63 (MH63)
Estrateji sekans
NGS li + HiFi li + CLR li + BioNano + Hi-C
Done:
ZS97: 8.34 Gb(~23x)HiFi li + 48.39 Gb (~131x) CLR li + 25 Gb (~69x) NGS + 2 selil BioNano Irys
MH63: 37.88 Gb (~103x) HiFi li + 48.97 Gb (~132x) CLR li + 28 Gb (~76x) NGS + 2 selil BioNano Irys
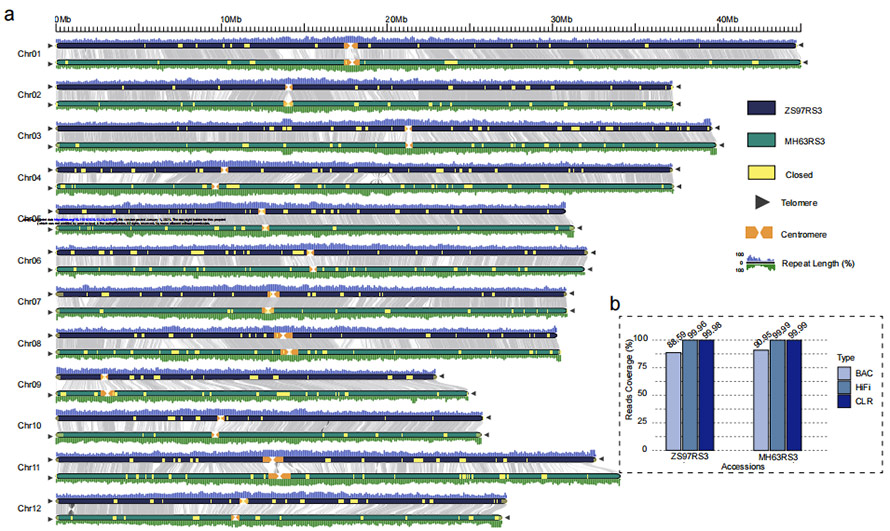
Figi 1 De jenòm diri san espas (MH63 ak ZS97)
2ndJenòm bannann2
Tit: Kwomozòm telomè-a-telomè nan bannann lè l sèvi avèk sekans nanopor
Doi:https://doi.org/10.1101/2021.04.16.440017
Lè afiche: 17 avril 2021.
Enstiti: Université Paris-Saclay, Frans
Materyèl
Double aploidMusa acuminatasppmalakensis(DH-Pahang)
Estrateji sekans ak done:
HiSeq2500 PE250 mòd + MinION/ PromethION (93Gb, ~ 200X)+ Kat optik (DLE-1+BspQ1)
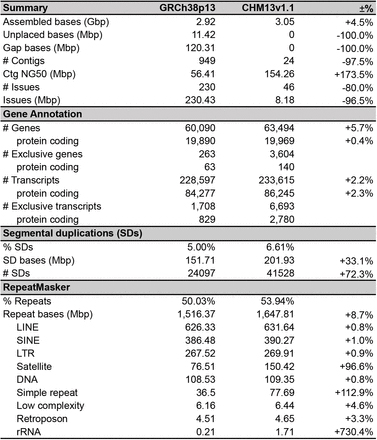
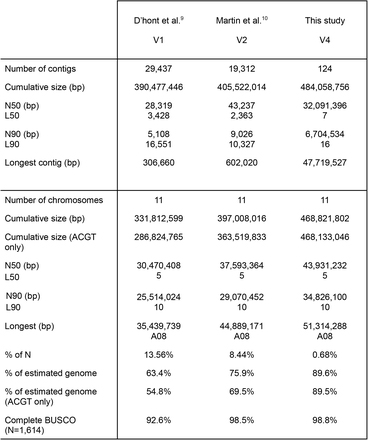
Tablo 1 Konparezon asanble genòm Musa acuminata (DH-Pahang).
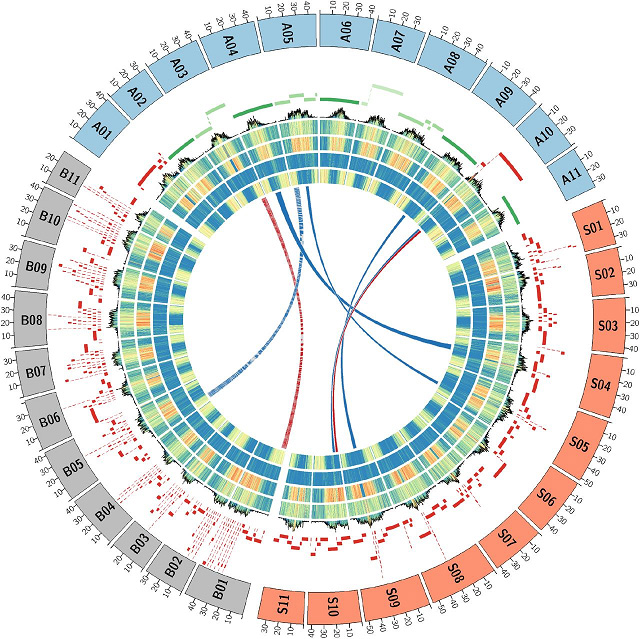
Figi 2 Musa genomes konparezon achitekti
3rdPhaeodactylum tricornutum genòm3
Tit: Telomere-a-telomere genome asanble nanP
haeodactylum tricornutum
Doi:https://doi.org/10.1101/2021.05.04.442596
Lè afiche: 04 me 2021
Enstiti: Western University, Kanada
Materyèl
Phaeodactylum tricornutum(Koleksyon Kilti Alg ak Protozoa CCAP 1055/1)
Estrateji sekans ak done:
1 Oxford Nanopore minION selil koule + yon 2 × 75 pè-fen mitan pwodiksyon NextSeq 550 kouri
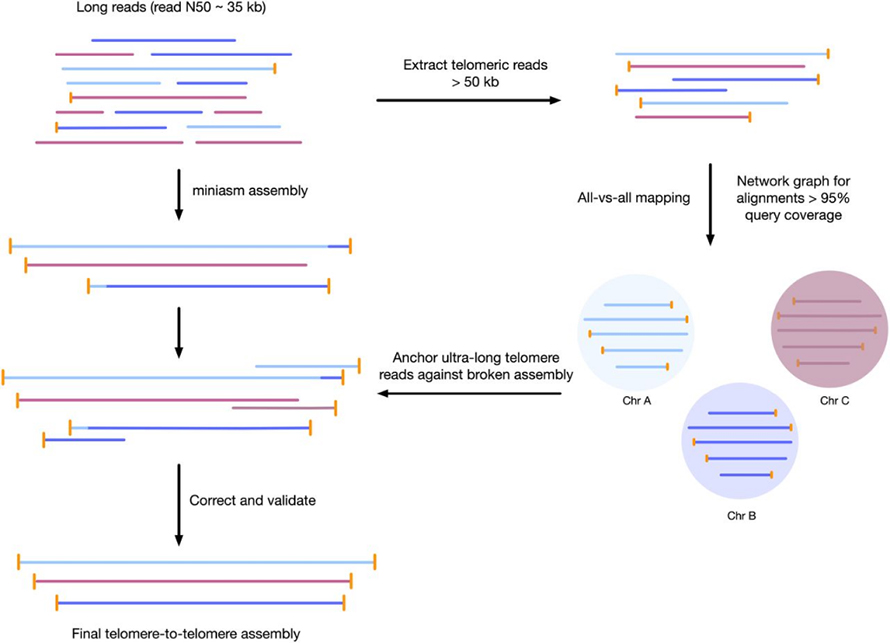
Figi 3 Workflow pou asanble telomere-a-telomere genome
4thJenòm CHM13 imen4
Tit: Sekans konplè yon genòm imen
Doi:https://doi.org/10.1101/2021.05.26.445798
Lè afiche: 27 me 2021
Enstiti: Enstiti Nasyonal Sante (NIH), USA
Materyèl: liy selilè CHM13
Estrateji sekans ak done:
30 × PacBio sikilè konsansis sekans (HiFi), 120 × Oxford Nanopore ultra-long sekans lekti, 100 × Illumina PCR-Free sekans (ILMN), 70 × Illumina / Arima Genomics Hi-C (Hi-C), kat optik BioNano, ak Strand-seq
Tablo 2 Konparezon GRCh38 ak T2T-CHM13 asanble genomic imen
Referans
1.Sergey Nurk et al.Sekans konplè yon genomic imen.bioRxiv 2021.05.26.445798;fè:https://doi.org/10.1101/2021.05.26.445798
2.Caroline Belser et al.Telomere-a-telomere gapless kwomozòm nan bannann lè l sèvi avèk sekans nanopor.bioRxiv 2021.04.16.440017;fè:https://doi.org/10.1101/2021.04.16.440017
3.Daniel J. Giguere et al.Telomere-a-telomere genomic asanble Phaeodactylum tricornutum.bioRxiv 2021.05.04.442596;fè:https://doi.org/10.1101/2021.05.04.442596
4.Jia-Ming Song et al.Asanble ak Validasyon de jenom referans ki pa gen diferans pou diri Xian/indica revele apèsi sou Achitekti Plant Centromere.bioRxiv 2020.12.24.424073;fè:https://doi.org/10.1101/2020.12.24.424073
Lè poste: Jan-06-2022

