Hi-C based Chromatin Interaction
Service Features
● Sequencing on Illumina NovaSeq with PE150.
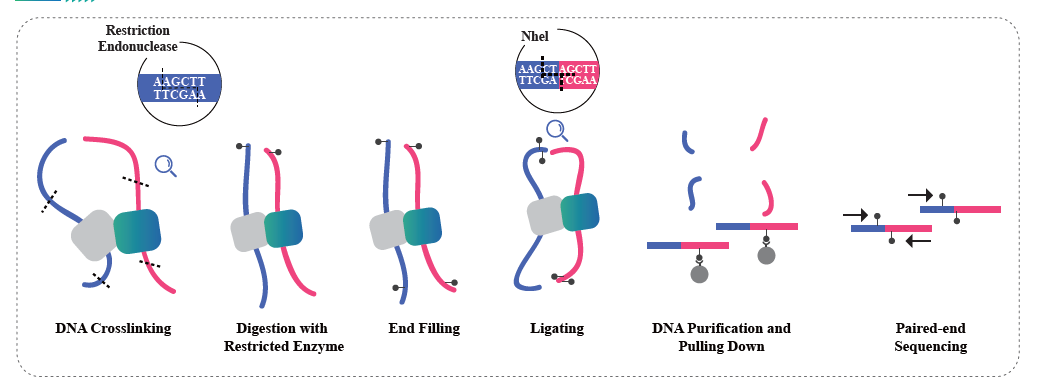
● Service requires tissue samples, instead of extracted nucleic acids, to cross-link with formaldehyde and conserve the DNA-protein interactions.
● The Hi-C experiment involves restriction and end repair of the sticky ends with biotin, followed by circularization of the resulting blunt ends while preserving the interactions. The DNA is then pulled down with streptavidin beads and purified for subsequent library preparation.
Service Advantages
● Optimal Restriction Enzyme Design: to ensure a high Hi-C efficiency on different species with up to 93% valid interaction pairs.
● Extensive Expertise and Publication Records: BMKGene has vast experience with >2000 Hi-C sequencing projects from 800 different species and various patents. Over 100 published cases with an accumulative impact factor of over 900.
● Highly-skilled Bioinformatics Team: with in-house patents and software copyrights for Hi-C experiments and data analysis and a self-developed visualization data software.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
● Comprehensive Annotation: we use multiple databases to functionally annotate the genes with identified variations and perform the corresponding enrichment analysis, providing insights on multiple research projects.
Service Specifications
|
Library |
Sequencing Strategy |
Recommended data output |
Hi-C Signal Resolution |
|
Hi-C library |
Illumina PE150 |
Chromatin loop: 150x TAD: 50x |
Chromatin Loop: 10Kb TAD: 40Kb |
Service Requirments
|
Sample type |
Required amount |
|
Animal tissue |
≥2g |
|
Whole Blood |
≥2mL |
|
Fungi |
≥1g |
|
Plant- young tissue |
1g/aliquot, 2-4 aliquotes recommended |
|
Cultured Cells |
≥1x107 |
Includes the Following Analysis:
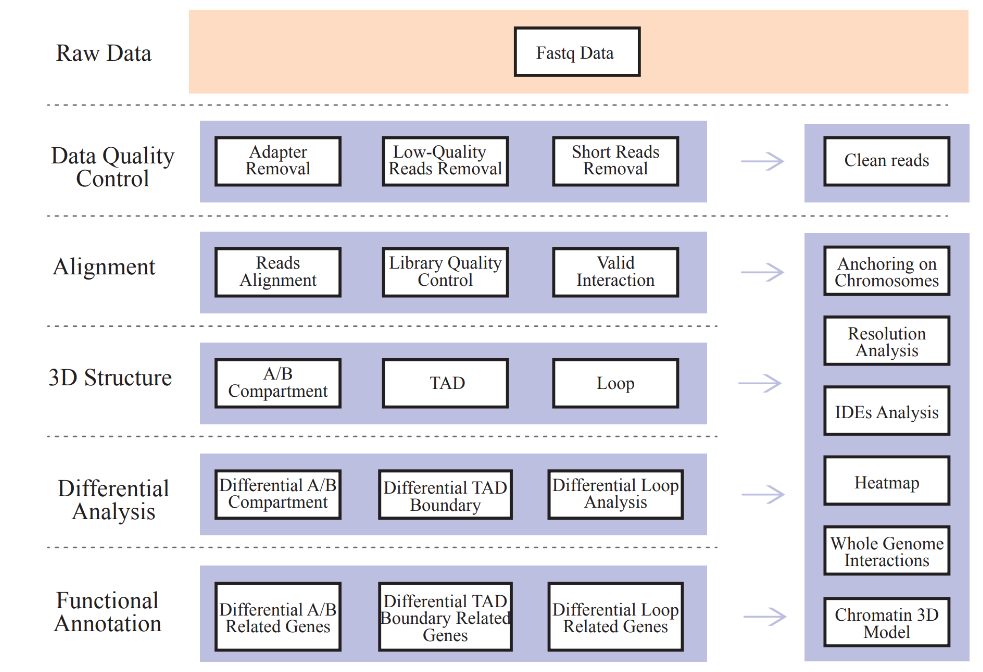
● Raw data QC;
● Mapping and Hi-C library QC: valid interaction pairs and Interaction Decay Exponents (IDEs);
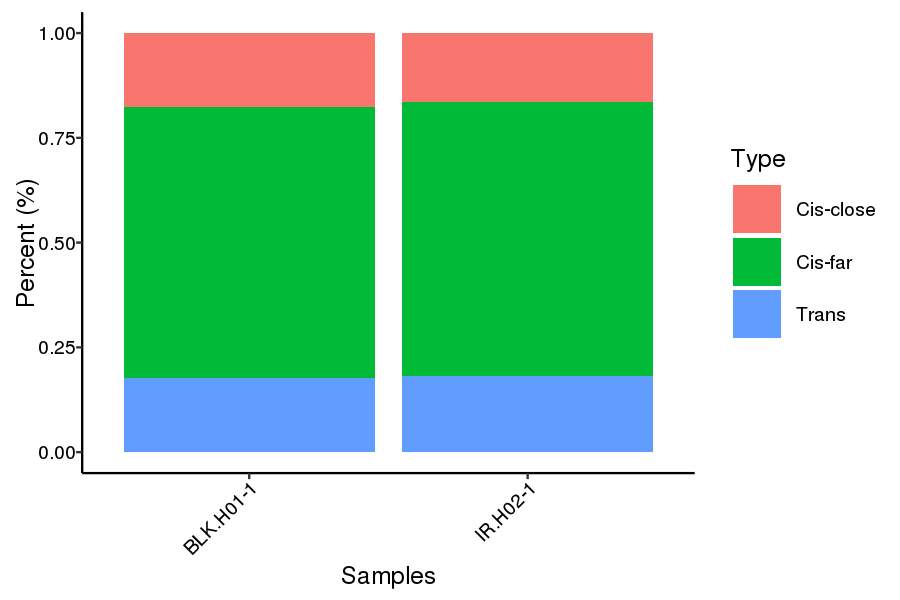
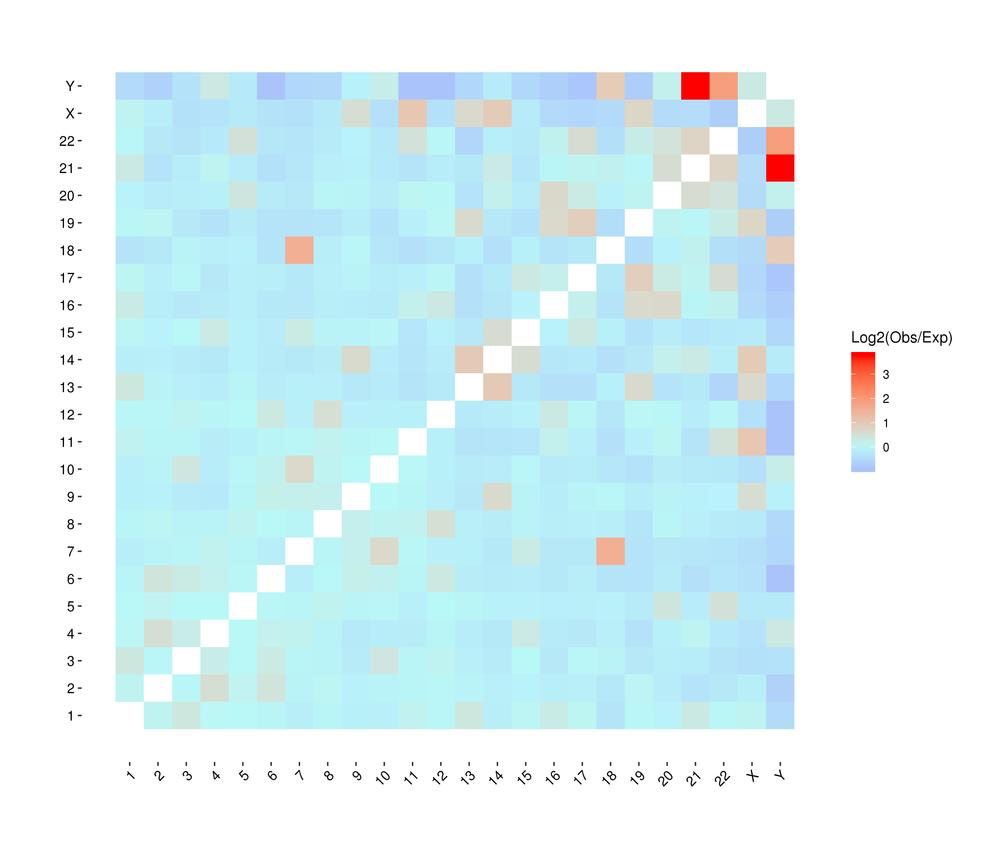
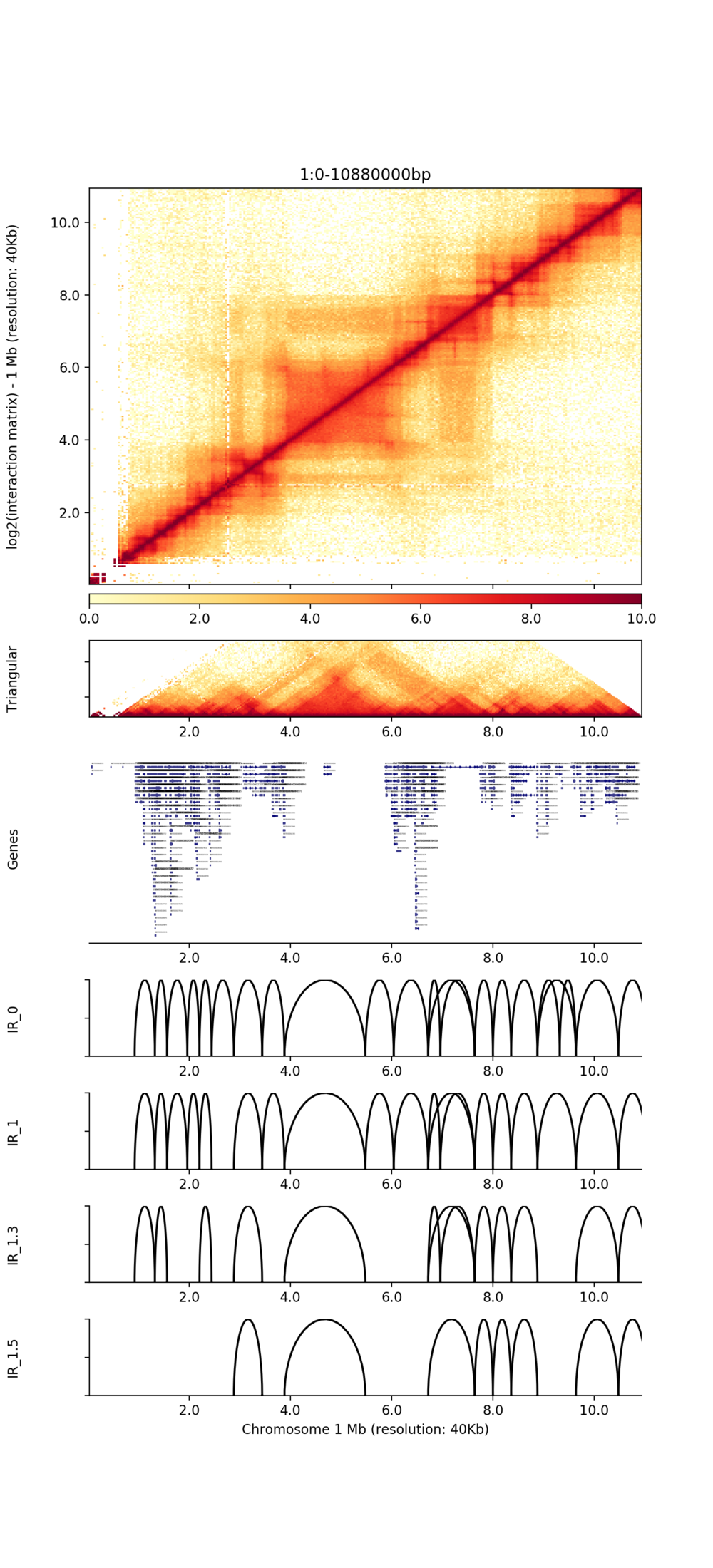
● Genome-wide Interaction Profiling: cis/trans analysis and Hi-C interaction map;
● Analysis of A/B compartment distribution;
● Identification of TADs and chromatin loops;
● Differential analysis on 3D chromatin structure elements among samples and corresponding functional annotation of associated genes.
Cis and trans proportion distribution
Heatmap of chromosomal interactions between samples
Genome-wide distribution of A/B compartments
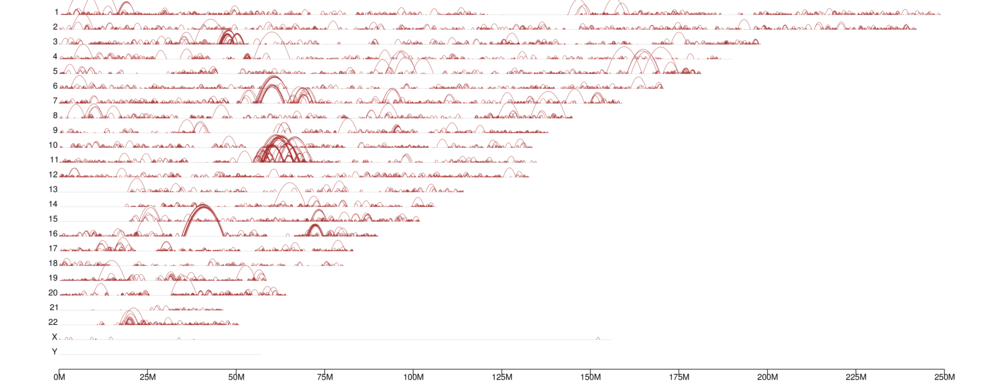
Genome-wide distribution of chromatin loops
Visualization of TADs
Explore the research advancements facilitated by BMKGene’s Hi-C sequencing services through a curated collection of publications.
Meng, T. et al. (2021) ‘A comparative integrated multi-omics analysis identifies CA2 as a novel target for chordoma’, Neuro-Oncology, 23(10), pp. 1709–1722. doi: 10.1093/NEUONC/NOAB156.
Xu, L. et al. (2021) ‘3D disorganization and rearrangement of genome provide insights into pathogenesis of NAFLD by integrated Hi-C, Nanopore, and RNA sequencing’, Acta Pharmaceutica Sinica B, 11(10), pp. 3150–3164. doi: 10.1016/J.APSB.2021.03.022.