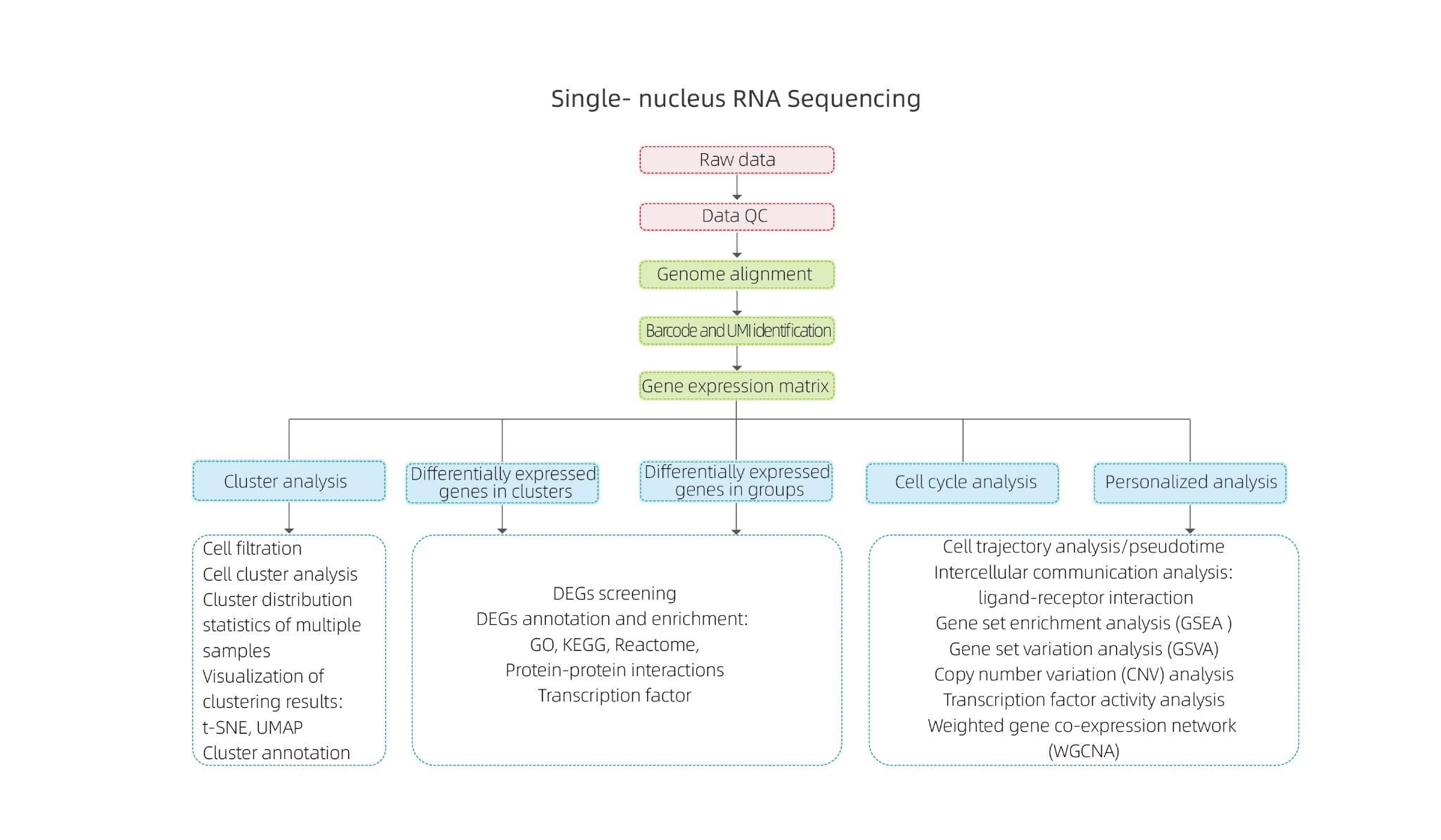
Hoʻokahi-nucleus RNA Sequencing
Kuʻuna loea
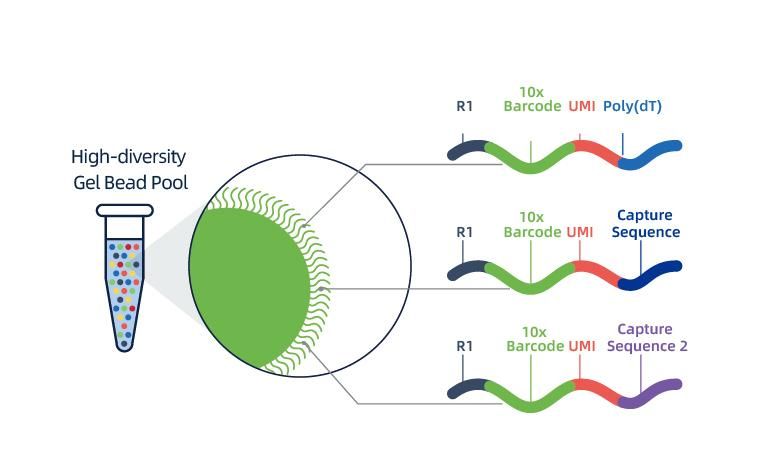
Loaʻa ka hoʻokaʻawale ʻana o ka nuclei e 10 × Genomics Chromium TM, nona ka ʻōnaehana microfluidics ʻewalu-channel me nā ala ʻelua.Ma kēia ʻōnaehana, ua hoʻopili ʻia kahi pahu gel me nā barcodes a me ka primer, nā enzymes a me kahi nucleus hoʻokahi i loko o ka kulu aila nui nanoliter, e hana ana i ka Gel Bead-in-Emulsion (GEM).Ke hoʻokumu ʻia ka GEM, hana ʻia ka cell lysis a me ka hoʻokuʻu ʻana i nā barcode i kēlā me kēia GEM.Hoʻololi ʻia ka mRNA i loko o nā molekala cDNA me 10x barcodes a me UMI, kahi e pili ana i ke kūkulu ʻana i ka hale waihona puke.
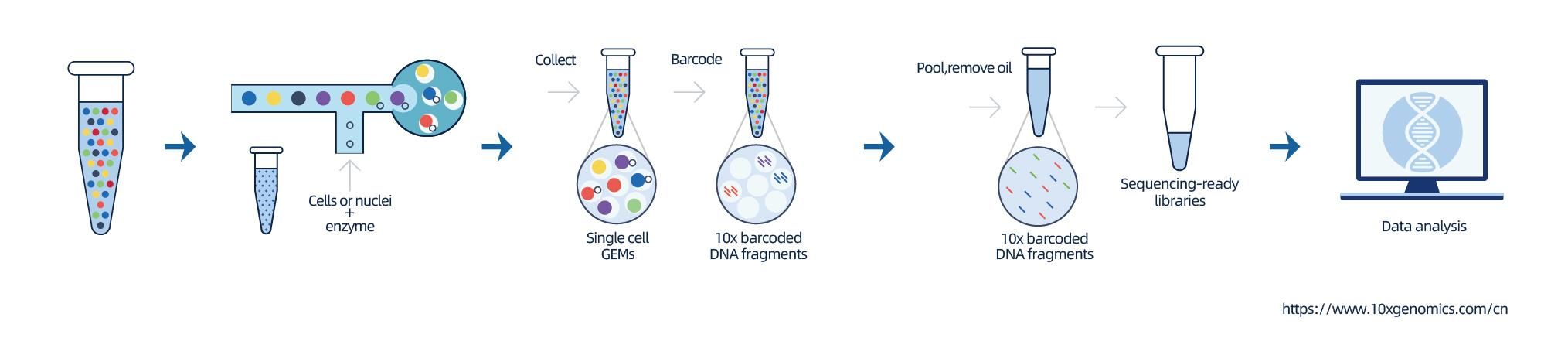
ʻAʻole kūpono ka ʻupena no ka hoʻomākaukau ʻana i ka hoʻopaneʻe ʻana o ke kelepona hoʻokahi
| Kelepona / Tissue | Ke kumu |
| Wehe hou i ke kino hau | ʻAʻole hiki ke loaʻa i nā hui hou a i mālama ʻia paha |
| ʻO ka ʻiʻo ʻiʻo, Megakaryocyte, Fat… | Nui loa ke anawaena o ke kelepona no ke komo ʻana i ka mea kani |
| ʻĀpaʻa… | ʻAʻole hiki ke hoʻokaʻawale i nā cell hoʻokahi |
| Kelepona neuron, lolo… | ʻOi aku ka maʻalahi, maʻalahi i ke koʻikoʻi, e hoʻololi i nā hopena sequencing |
| Pancreas, Thyroid… | Loaʻa i nā enzymes endogenous, e pili ana i ka hana ʻana o ka hoʻokuʻu ʻana o ke kelepona hoʻokahi |
Single-nucleus vs Single-cell
| Hoʻokahi-nucleus | pakahi-kele |
| ʻO ke anawaena kelepona palena ʻole | Ke anawaena pūnaewele: 10-40 μm |
| Hiki i nā mea ke lilo i mea hau | ʻO ka mea pono he kikokiko hou |
| ʻO ke koʻikoʻi haʻahaʻa o nā pūnaewele hau | Hiki i ka hoʻohana ʻana i ka enzyme ke hoʻoulu i ke koʻikoʻi o ka cell |
| ʻAʻole pono e wehe ʻia ke koko ʻulaʻula | Pono e wehe ʻia ke koko ʻulaʻula |
| Hōʻike ka Nuclear i ka bioinformation | Hōʻike ke kelepona holoʻokoʻa i ka bioinformation |
Nā kikoʻī lawelawe
| Hale Waihona Puke | Hoʻolālā kaʻina | Volume ʻIkepili | Nā Koina Laʻana | Kiʻi |
| 10× Genomics hoʻokahi-nuclei waihona | 10x Genomics -Illumina PE150 | 100,000 heluhelu/cell approx.100-200 Gb | Helu kelepona: >2× 105 Cell conc.ma 700-1,200 cell / μL | ≥ 200 mg |
No nā kikoʻī hou aku e pili ana i ke alakaʻi hoʻomākaukau laʻana a me ke kahe hana lawelawe, e ʻoluʻolu e kamaʻilio me aloea BMKGENE
Holo Hana Hana

Hoʻolālā hoʻokolohua

Hāʻawi laʻana
Kaʻawale nuclei

Kūkulu hale waihona puke

Ke kaʻina ʻana

ʻIkepili ʻikepili

Nā lawelawe ma hope o ke kūʻai aku
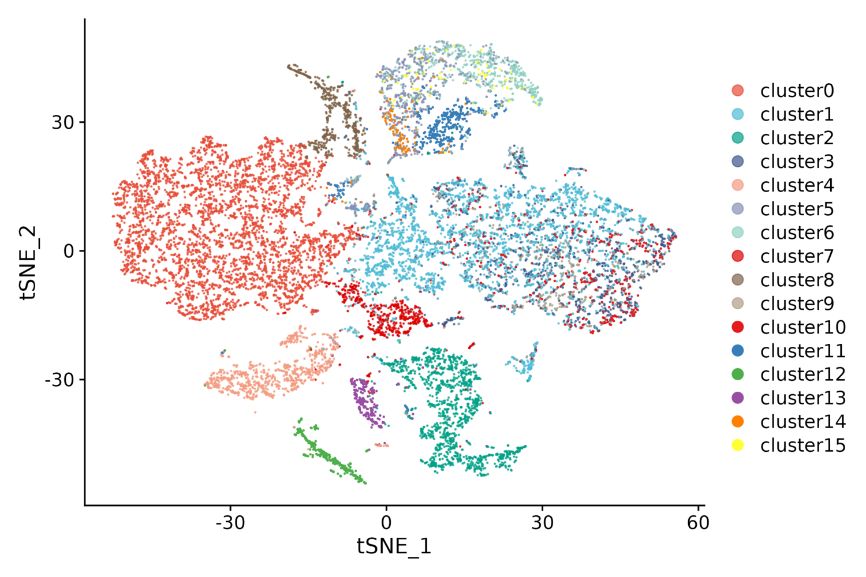
1.Spot clustering
2. Hōʻike hōʻailona abundance clustering heatmap
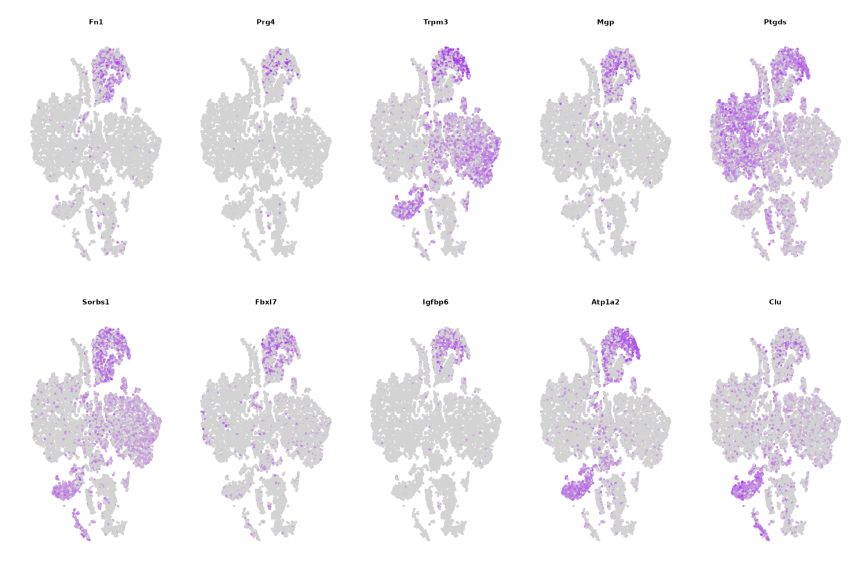
 3.Maker gene distribution in different clusters
3.Maker gene distribution in different clusters
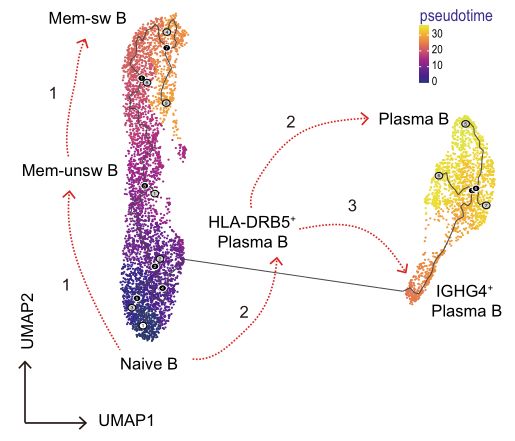
4.Cell trajectory analysis/pseudotime