
Nā Genetics Evolutionary
Pono lawelawe
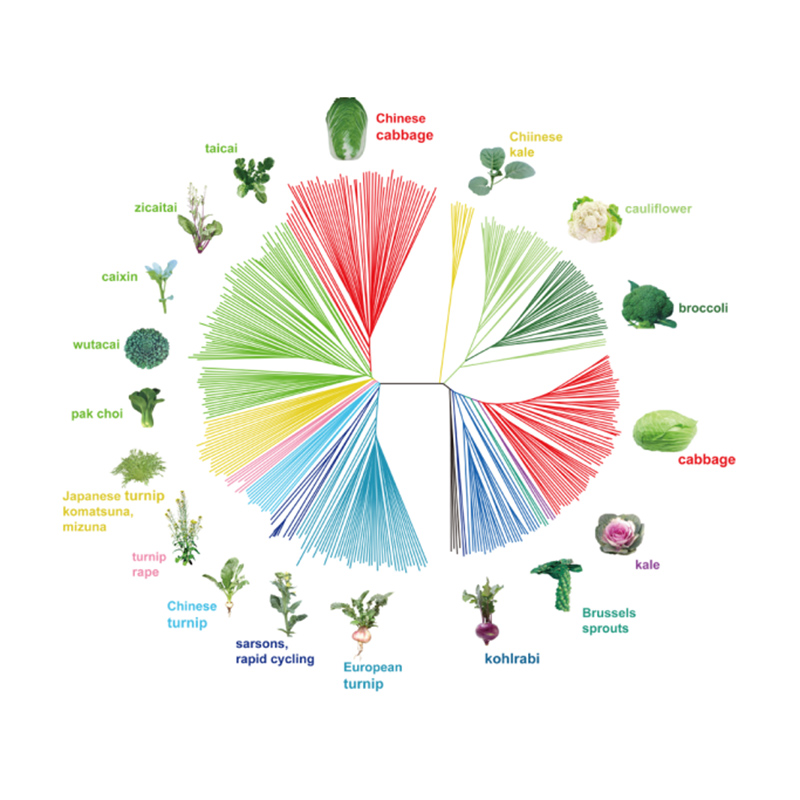
Takagi et al.,Ka puke pai mea kanu, 2013
● Ke koho ʻana i ka manawa like ʻole a me ka wikiwiki e pili ana i nā ʻano like ʻole ma ka pae nucleotide a me nā waikawa amino.
● Ka hōʻike ʻana i ka pilina pili phylogenetic hilinaʻi ma waena o nā ʻano me ka mana liʻiliʻi o ka hoʻololi hoʻololi ʻana a me ka hoʻololi like.
● Ke kūkulu ʻana i nā loulou ma waena o nā hoʻololi genetic a me nā phenotypes e wehe i nā genes pili i ke ʻano
● Ka helu ʻana i ka ʻokoʻa ʻokoʻa, e hōʻike ana i ka hiki ke hoʻololi o nā ʻano
● ʻOi aku ka wikiwiki o ka huli ʻana
● ʻIke nui: Ua hōʻiliʻili ʻo BMK i ka ʻike nui i ka heluna kanaka a me nā papahana pili i ka evolutionary no nā makahiki he 12, e uhi ana i nā haneli haneli, etc.
Nā kikoʻī lawelawe
Mea Hana:
ʻO ka maʻamau, ʻoi aku ka liʻiliʻi o ʻekolu mau sub-populations (e laʻa i nā ʻano subspecies a i ʻole nā kauā).Pono ka heluna kanaka ma lalo o 10 mau kānaka (Nā mea kanu >15, hiki ke hoʻemi ʻia no nā ʻano laha ʻole).
Hoʻolālā kaʻina hana:
* Hiki ke hoʻohana ʻia ʻo WGS no nā ʻano me nā genome kuhikuhi kiʻekiʻe, ʻoiai pili ʻo SLAF-Seq i nā ʻano me ka ʻole a i ʻole ka genome kuhikuhi, a i ʻole ka genome kuhikuhi maikaʻi ʻole.
| Pili i ka nui genome | WGS | SLAF-Hui (×10,000) |
| ≤ 500 Mb | 10×/hoʻokahi | ʻOi aku ka manaʻo o WGS |
| 500 Mb - 1 Gb | 10 | |
| 1 Gb - 2 Gb | 20 | |
| ≥2 Gb | 30 |
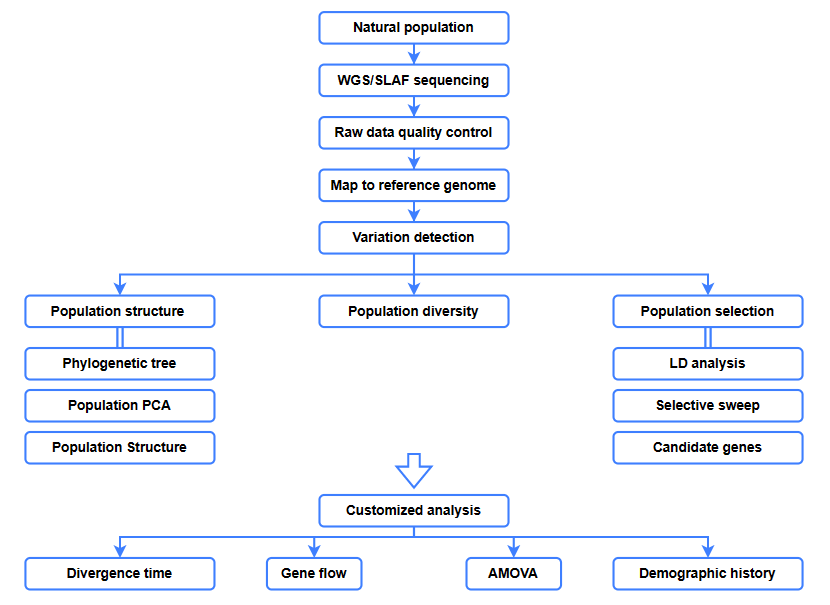
Nā loiloi bioinformatics
● Ke kālailai hoʻololi
● Holoi koho
● Ke kahe ʻana o Gene
● mōʻaukala kanaka
● Ka manawa like ʻole
Nā Koina Laʻana a me ka Hoʻouna ʻana
Nā Koina Laʻana:
| ʻAnoʻano | Kiʻi | WGS-NGS | SLAF |
| Holoholona
| ʻiʻo visceral |
0.5-1g
|
0.5g
|
| ʻiʻo ʻiʻo | |||
| Koko momona | 1.5mL
| 1.5mL
| |
| Koko moa/i'a | |||
| Meakanu
| Lau Hou | 1~2g | 0.5-1g |
| Petal/Stem | |||
| Aʻa/Hua | |||
| Nā pūnaewele | Cell moʻomeheu |
| gDNA | Hoʻopaʻa ʻana | Ka nui (ʻā) | OD260/OD280 |
| SLAF | ≥35 | ≥1.6 | 1.6-2.5 |
| WGS-NGS | ≥1 | ≥0.1 | - |
Holo Hana Hana

Hoʻolālā hoʻokolohua

Hāʻawi laʻana

Kūkulu hale waihona puke

Ke kaʻina ʻana

ʻIkepili ʻikepili

Nā lawelawe ma hope o ke kūʻai aku
*ʻO nā hopena demo i hōʻike ʻia ma ʻaneʻi mai nā genomes i paʻi ʻia me BMKGENE
1. Loaʻa i ka loiloi Evolution ke kūkulu ʻana i ka lāʻau phylogenetic, ka hoʻonohonoho ʻana o ka heluna kanaka a me ka PCA ma muli o nā ʻano like ʻole.
Hōʻike ka lāʻau phylogenetic i ka pili ʻauhau a me ka evolutionary ma waena o nā ʻano me nā kūpuna maʻamau.
Manaʻo ʻo PCA e ʻike i ka pili kokoke i waena o nā sub-populations.
Hōʻike ka ʻōnaehana heluna i ka hiki ʻana mai o nā sub-population ʻokoʻa ma ke ʻano o nā alapine allele.
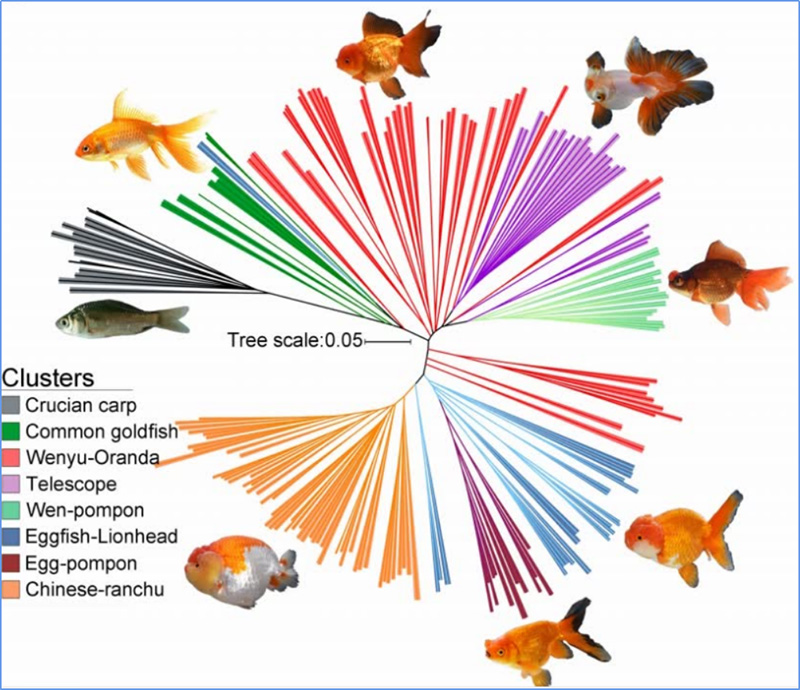
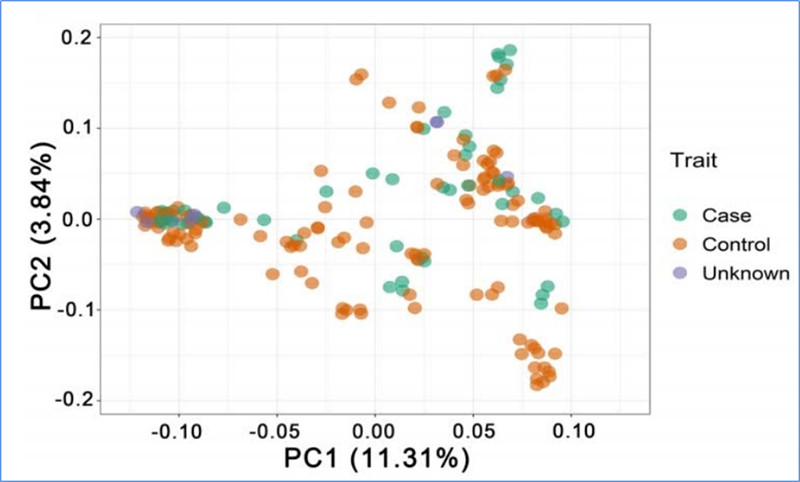
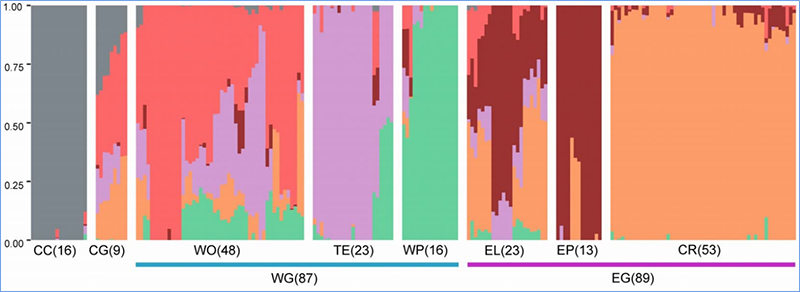
ʻO Chen, et.al.,PNAS, 2020
2. Holoi koho
ʻO ka sweep koho e pili ana i kahi kaʻina e koho ʻia ai kahi pūnaewele maikaʻi a hoʻonui ʻia nā alapine o nā pūnaewele kūʻokoʻa i hoʻopili ʻia a hoʻemi ʻia nā pūnaewele i hoʻopaʻa ʻole ʻia, e hopena i ka hōʻemi ʻana o ka paena.
Hoʻohana ʻia ka ʻike ʻana i ka genome-ākea ma nā wahi kahiki koho ʻia ma ka helu ʻana i ka helu helu helu helu heluna kanaka (π,Fst, Tajima's D) o nā SNP a pau i loko o kahi puka aniani (100 Kb) ma kekahi pae (10 Kb).
ʻokoʻa nucleotide(π)
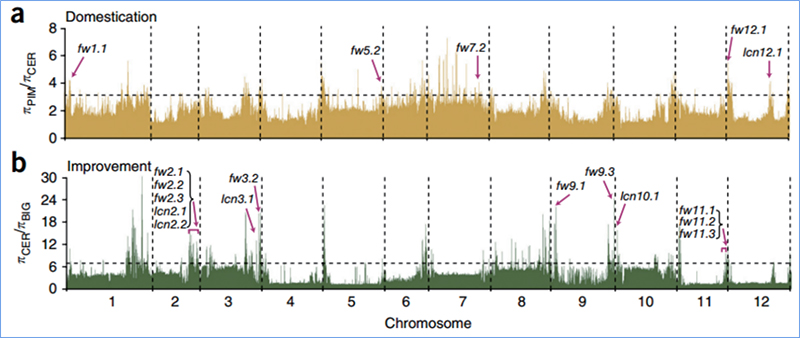
ʻO D
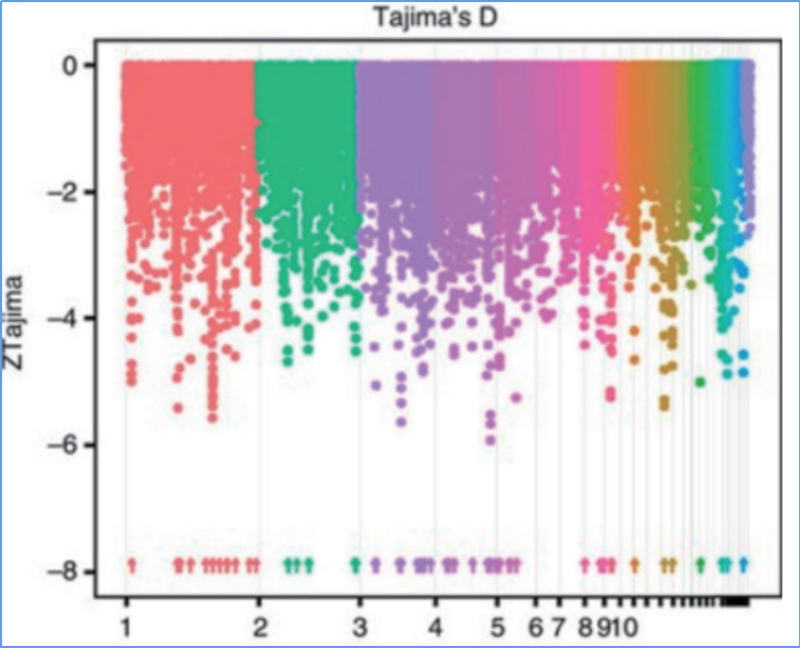
Helu kuhikuhi (Fst)
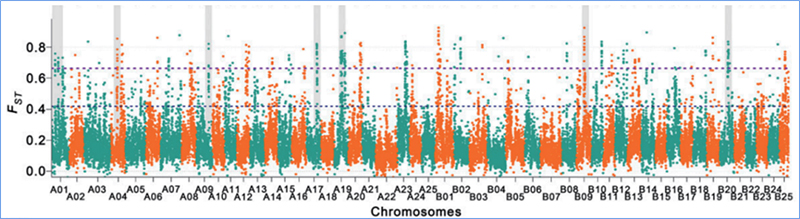
ʻO Wu, et.al.,Mea Molekula, 2018
3.Gene Kahe
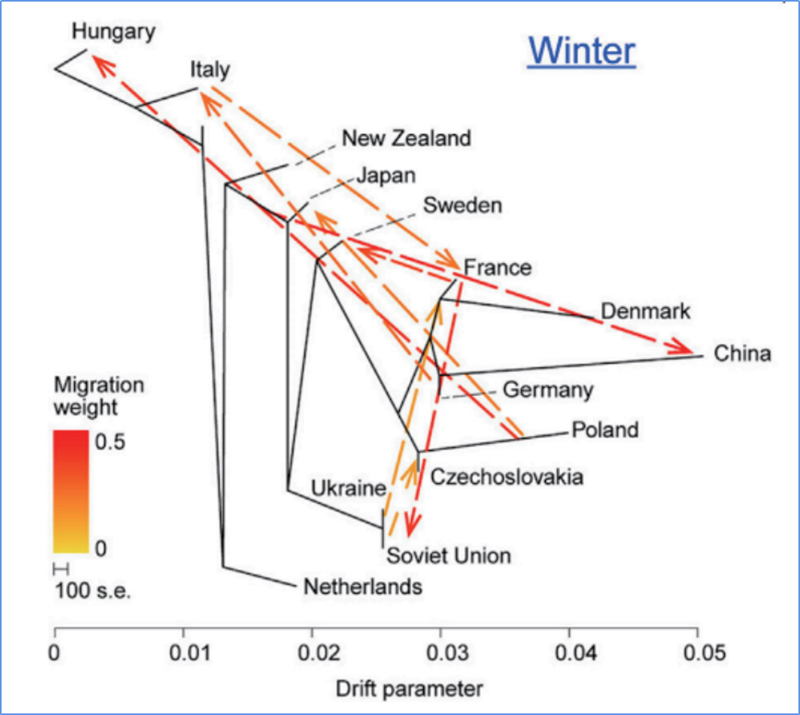
ʻO Wu, et.al.,Mea Molekula, 2018
4.Ka moʻolelo demographic
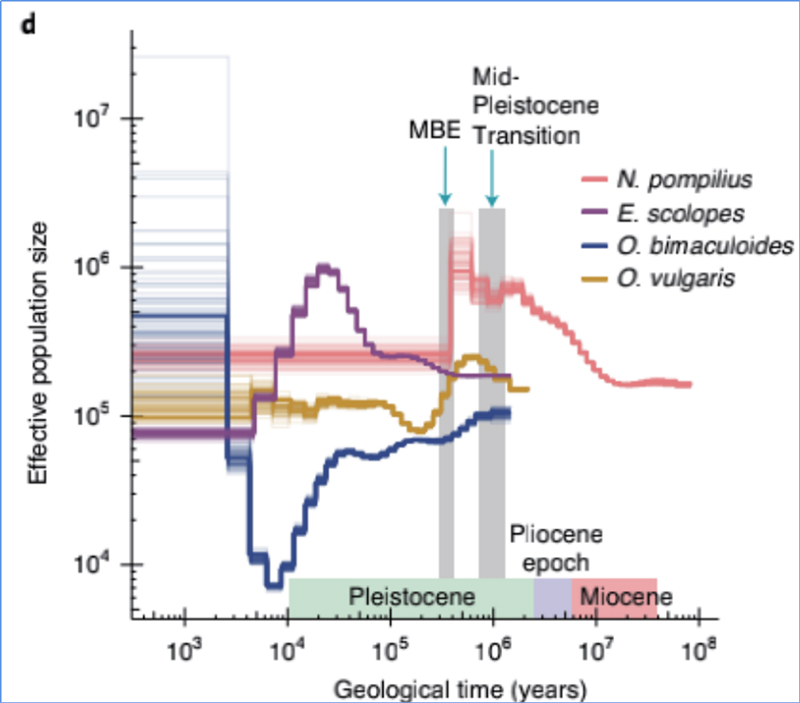
ʻO Zhang, et.al.,Nature Ecology & Evolution, 2021
5.Divergence manawa
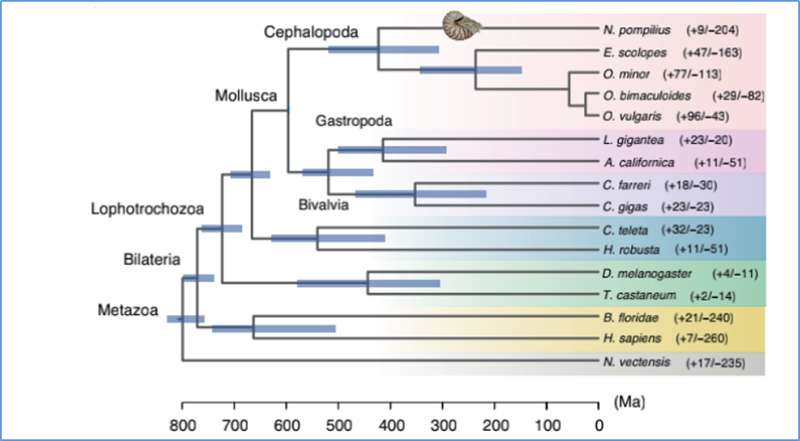
ʻO Zhang, et.al.,Nature Ecology & Evolution, 2021
Hihia BMK
Hāʻawi ka palapala ʻāina hoʻololi genomic i nā ʻike i ke kumu moʻo o ke koho ʻana o Spring Chinese Cabbage (Brassica rapa ssp. Pekinensis)
Paʻi ʻia: Mea Molekula, 2018
Hoʻolālā kaʻina hana:
Ke hoʻomau nei: ka hohonu o ke kaʻina: 10 ×
Nā hopena koʻikoʻi
Ma kēia noiʻi ʻana, ua hana ʻia nā kāpeti Pākē 194 no ka hoʻonohonoho hou ʻana me ka hohonu awelika o 10x, a loaʻa iā 1,208,499 SNPs a me 416,070 InDels.Hōʻike ka loiloi phylogenetic ma kēia mau laina 194 e hiki ke hoʻokaʻawale ʻia kēia mau laina i ʻekolu ecotypes, puna, kauwela a me ka hāʻule.Eia hou, ua hōʻike ʻia ka hoʻonohonoho ʻana o ka heluna kanaka a me ka nānā ʻana i ka PCA no ke kāpeti ʻālau ma Shandong, Kina.Ua hoʻolauna ʻia kēia mau mea i Korea a me Iapana, ua hele ʻia me nā laina kūloko a ua hoʻihoʻi ʻia kekahi mau ʻano bolting hope o lākou i Kina a ua lilo i kāpeti Pākē Spring.
Ua hōʻike ʻia he 23 genomic loci i ka mākaʻikaʻi ʻana o Genome-ākea i nā kāpeti Kina punawai a me nā kāpeti hāʻule i koho ʻia, ʻelua o ia mau mea i uhi ʻia me ka ʻāina hoʻomalu manawa bolting e pili ana i ka QTL-mapping.Ua ʻike ʻia kēia mau ʻāpana ʻelua i loko o nā genes nui e hoʻoponopono i ka pua, BrVIN3.1 a me BrFLC1.Ua hōʻoia hou ʻia kēia mau genes ʻelua e komo i ka manawa bolting e ka transcriptome study a me nā hoʻokolohua transgenic.
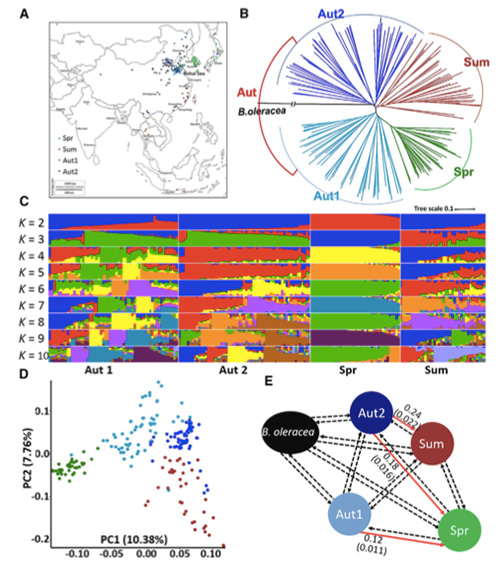 ʻO ka nānā ʻana i ka hoʻokumu ʻana o ka lehulehu ma nā kāpeti Pākē | 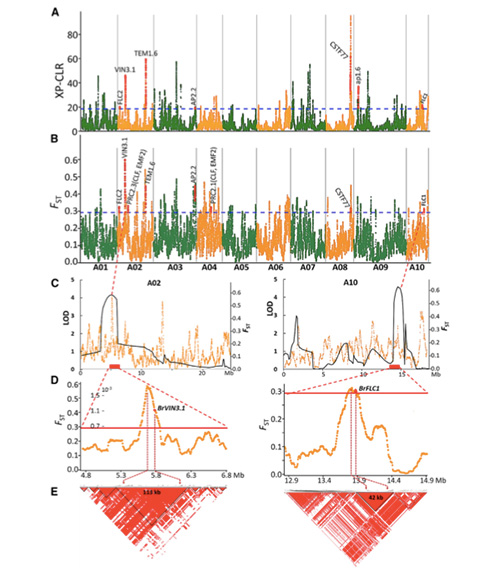 ʻIke genetic e pili ana i ke koho kāpeti Kina |
Tongbing, et al.“Hāʻawi ʻia kahi palapala ʻāina hoʻololi Genomic i nā ʻike i ke kumu hoʻohālikelike o ke koho ʻana i ke kāpeti Pākē puna (Brassica rapa ssp.pekinensis).Nā mea kanu molekula,11(2018):1360-1376.















