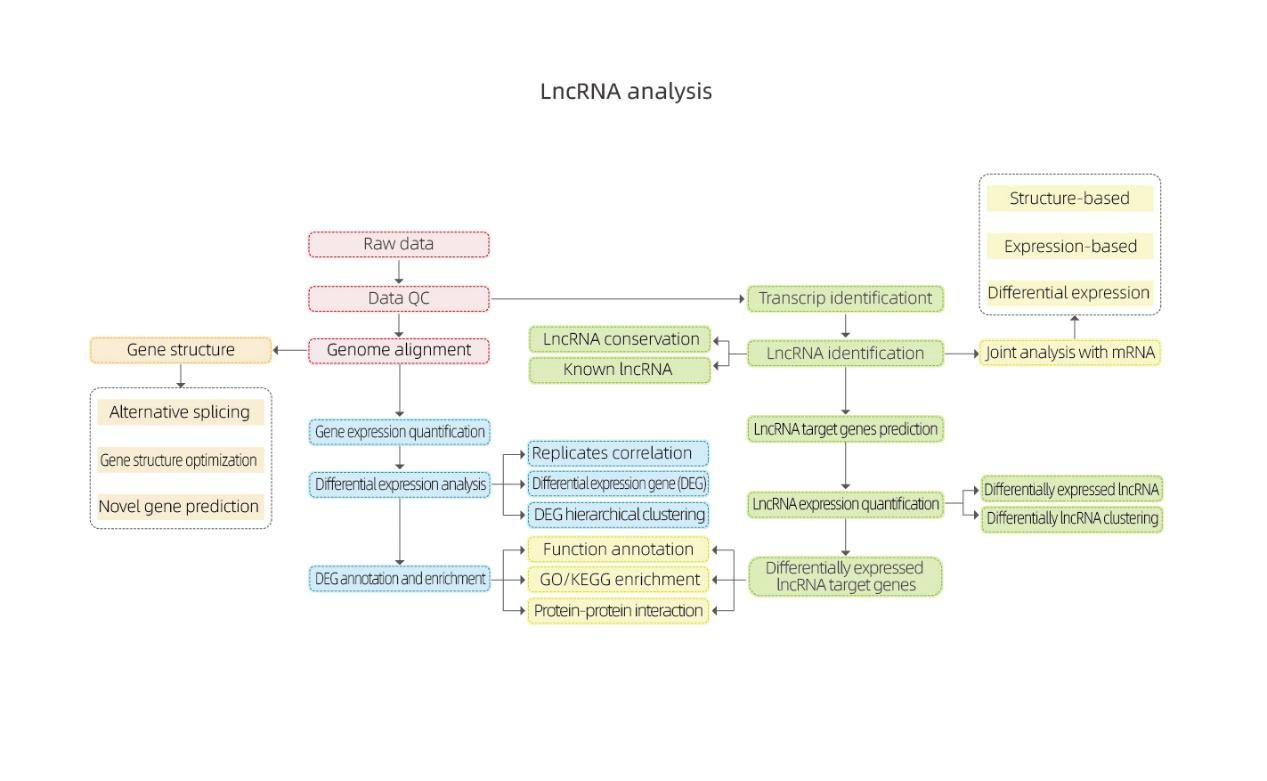
Dogon jerin abubuwan da ba codeing-Ilumina
Amfanin Sabis
● Amfanin Sabis
● Kwayoyin salula da takamaiman nama
Takaitaccen mataki yana bayyanawa da gabatar da canjin magana mai ƙarfi
● Madaidaicin tsarin lokacin magana da sarari
● Binciken haɗin gwiwa tare da bayanan mRNA.
● Isar da sakamako na tushen BMKCloud: Ma'adinin bayanai na musamman da ake samu akan dandamali.
● Sabis na tallace-tallace yana aiki na tsawon watanni 3 bayan kammala aikin
Samfuran Bukatun da Bayarwa
| Laburare | Dandalin | Bayanan da aka ba da shawarar | Bayanan Bayani na QC |
| raguwar rRNA | Farashin PE150 | 10 gb | Q30≥85% |
| Conc.(ng/μl) | Adadin (μg) | Tsafta | Mutunci |
| ≥ 100 | ≥ 0.5 | OD260/280=1.7-2.5 OD260/230=0.5-2.5 Iyakance ko babu furotin ko gurɓataccen DNA da aka nuna akan gel. | Don tsire-tsire: RIN≥6.5; Na dabbobi: RIN≥7.0; 5.0≥28S/18S≥1.0; iyakance ko babu hawan tushe |
Nucleotides:
Nama: Nauyi (bushe): ≥1 g
* Don nama ƙasa da 5 MG, muna ba da shawarar aika samfurin nama daskararre (a cikin ruwa nitrogen).
Dakatar da salula: Ƙididdiga ta salula = 3×107
*Muna ba da shawarar jigilar daskararrun cell lysate.Idan wannan tantanin halitta ya ƙidaya ƙasa da 5 × 105, walƙiya daskararre a cikin ruwa nitrogen ana shawarar.
Samfuran jini:
PA×geneBloodRNATube;
6mLTRIzol da 2ml jini (TRIzol:Blood=3:1)
Isar da Samfurin Nasiha
Kwantena: 2 ml bututu centrifuge (Ba a ba da shawarar foil tin ba)
Alamar samfur: Ƙungiya+ kwafi misali A1, A2, A3;B1, B2, B3......
Jirgin ruwa:
1.Dry-ice: Ana buƙatar samfurori a cikin jaka kuma a binne su a bushe-kankara.
2.RNAstable tubes: Ana iya bushe samfuran RNA a cikin bututun daidaitawar RNA (misali RNAstable®) kuma a tura su cikin zafin jiki.
Gudun Aikin Sabis

Gwajin ƙira

Samfurin bayarwa

RNA cirewa

Gina ɗakin karatu

Jeri

Binciken bayanai

Bayan-sayar da sabis
Bioinformatics
1.LncRNA rarrabawa
LncRNA da softwares guda huɗu da ke sama suka annabta an rarraba su zuwa rukuni 4: lincRNA, anti-sense-LncRNA, intronic-LncRNA;hankali-LncRNA.An nuna rarrabuwar LncRNA a cikin tarihin da ke ƙasa.
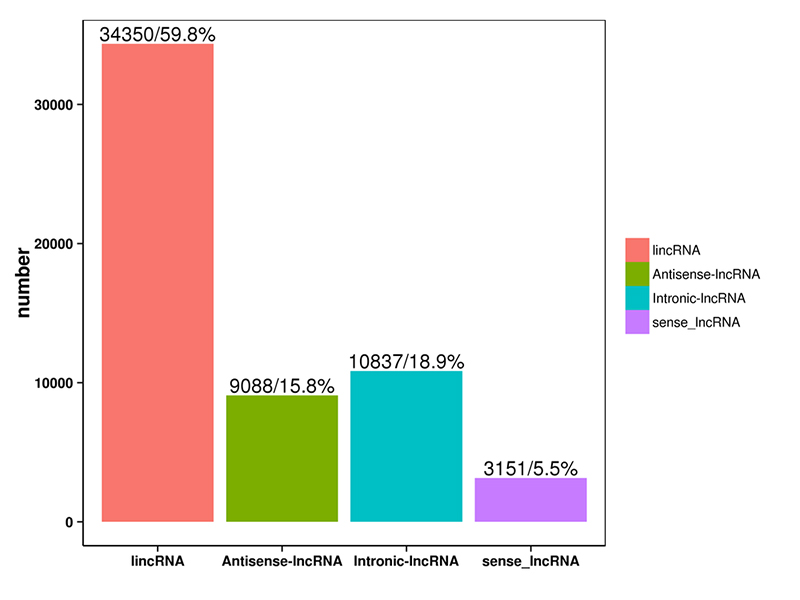
Rarraba LncRNA
2.Cis-manufa genes na DE-lncRNA inrichment analysis
An yi amfani da ClusterProfiler a cikin bincike na haɓaka GO akan kwayoyin halittar cis-cis na lncRNA (DE-lncRNA) da aka bayyana daban-daban, dangane da tsarin nazarin halittu, ayyukan ƙwayoyin cuta da sassan salula.Binciken haɓakar GO tsari ne don gano ƙa'idodin GO da aka ba da umarni sosai idan aka kwatanta da dukkan kwayoyin halitta.An gabatar da sharuɗɗan da aka wadatar a cikin histogram, ginshiƙi na kumfa, da sauransu kamar yadda aka nuna a ƙasa.
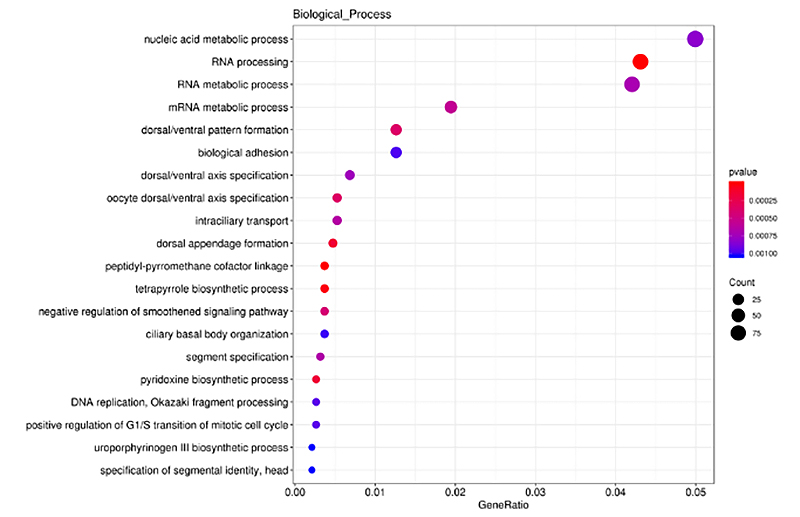 Kwayoyin Cis da aka yi niyya na DE-lncRNA nazarin haɓaka haɓaka - ginshiƙi
Kwayoyin Cis da aka yi niyya na DE-lncRNA nazarin haɓaka haɓaka - ginshiƙi
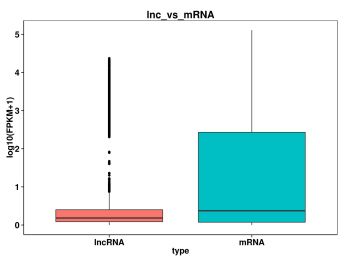
3. Ta hanyar kwatanta tsayi, lambar exon, ORF da adadin magana na mRNA da lncRNA, za mu iya fahimtar bambance-bambance a cikin tsari, jeri da sauransu a tsakanin su, da kuma tabbatar da ko littafin lncRNA da mu ya annabta ya dace da ainihin halaye.
BMK Case
Bayanan bayanan lncRNA da aka lalata a cikin huhu na linzamin kwamfuta adenocarcinomas tare da maye gurbin KRAS-G12D da P53 knockout
Buga:Jaridar Cellular and Molecular Medicine,2019
Dabarun jeri
Illumina
Tarin samfurin
Kwayoyin NONMMUT015812-knockdown KP (shRNA-2) da ƙwayoyin cuta mara kyau (sh-Scr) an sami su a ranar 6 na takamaiman kamuwa da cuta.
Sakamako mai mahimmanci
Wannan binciken yana bincika lncRNAs da aka bayyana a cikin huhu na linzamin kwamfuta adenocarcinoma tare da bugun P53 da maye gurbin KrasG12D.
1.6424 lncRNAs an bayyana daban-daban (≥ sau biyu sau biyu, P <0.05).
2.A cikin duk 210 lncRNAs (FC≥8), 11 lncRNAs' furcin P53 ne ya tsara shi, 33 lncRNAs ta KRAS da 13 lncRNAs ta hypoxia a cikin ƙananan ƙwayoyin KP na farko, bi da bi.
3.NONMMUT015812, wanda aka daidaita shi sosai a cikin huhu huhu adenocarcinoma kuma an tsara shi ta hanyar sake bayyana P53, don nazarin aikin salula.
4.Knockdown na NONMMUT015812 ta shRNAs sun rage yaduwa da damar ƙaura na ƙwayoyin KP.NONMMUT015812 ya kasance mai yuwuwar oncogene.
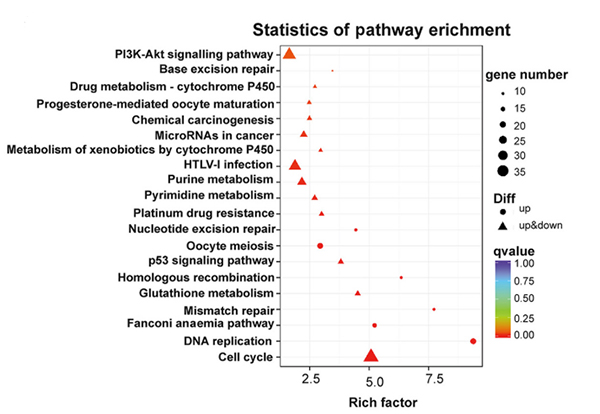 Binciken hanyar KEGG na nau'ikan jinsin da aka bayyana daban-daban a cikin sel NONMMUT015812-knockdown KP | 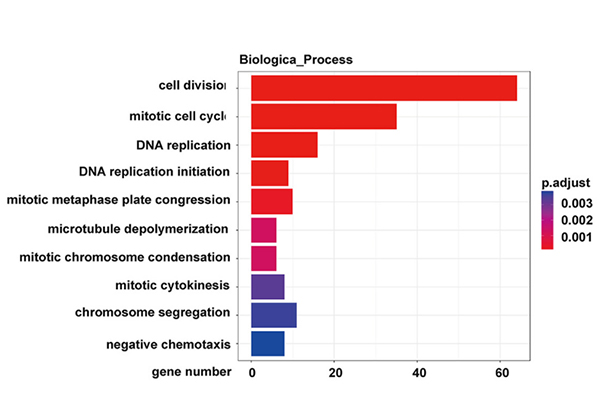 Binciken Gene Ontology na nau'ikan jinsin da aka bayyana daban-daban a cikin sel NONMMUT015812-knockdown KP |
Magana
Bayanan bayanan lncRNA da aka lalata a cikin huhu na linzamin kwamfuta adenocarcinomas tare da maye gurbin KRAS-G12D da P53 knockout[J].Jaridar Cellular and Molecular Medicine, 2019, 23(10).DOI: 10.1111/jcm.14584