De novo Fungal Genome Assembly
Service Features
With three possible options to choose from depending on the desired degree of genome completeness:
● Draft Genome Option: Short-read sequencing with Illumina NovaSeq PE150.
● Fungal Fine Genome Option:
Genome Survey: Illumina NovaSeq PE150.
Genome Assembly: PacBio Revio (HiFi reads) or Nanopore Promethion P48.
● Chromosome-level Fungal Genome:
Genome Survey: Illumina NovaSeq PE150.
Genome Assembly: PacBio Revio (HiFi reads) or Nanopore Promethion P48.
Contig anchoring with Hi-C assembly.
Service Advantages
● Multiple Sequencing Strategies Available: For different research goals and requirements of genome completeness
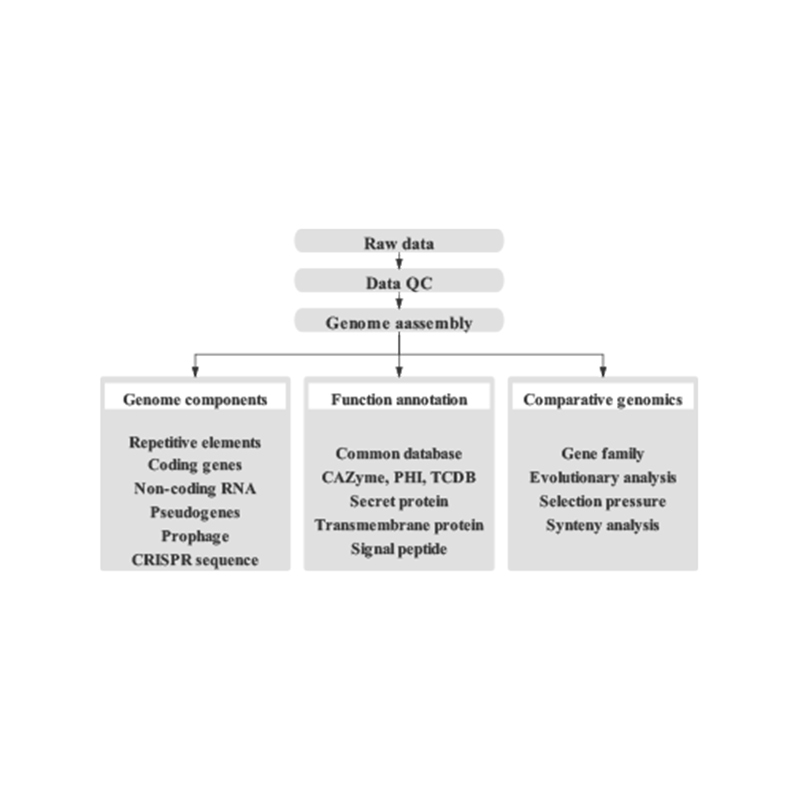
● Complete Bioinformatics Workflow: This includes genome assembly and the prediction of multiple genomic elements, functional gene annotation, and contig anchoring.
● Extensive Expertise: With over 10,000 microbial genomes assembled, we bring over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Service Specifications
|
Service |
Sequencing Strategy |
Quality Control |
|
Draft Genome |
Illumina PE150 100x |
Q30≥85% |
|
Fine Genome |
Genome survey: Illumina PE150 50 x Assembly: PacBio HiFi 30x or Nanopore 100x |
contig N50 ≥1Mb(Pacbio Unicellular) contig N50 ≥2Mb(ONT Unicellular) contig N50 ≥500kb(Others) |
|
Chromosome-level genome |
Genome survey: Illumina PE150 50 x Assembly: PacBio HiFi 30x or Nanopore 100x Hi-C Assembly 100x |
Contig anchoring ratio>90%
|
Service Requirments
|
Concentration (ng/µL) |
Total amount (µg) |
Volume (µL) |
OD260/280 |
OD260/230 |
|
|
PacBio |
≥20 |
≥2 |
≥20 |
1.7-2.2 |
≥1.6 |
|
Nanopore |
≥40 |
≥2 |
≥20 |
1.7-2.2 |
≥1 |
|
Illumina |
≥1 |
≥0.06 |
≥20 |
1.7-2.2 |
- |
Unicellular Fungus: ≥3.5x1010 cells
Macro Fungus: ≥10 g
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
Includes the Following Analysis:
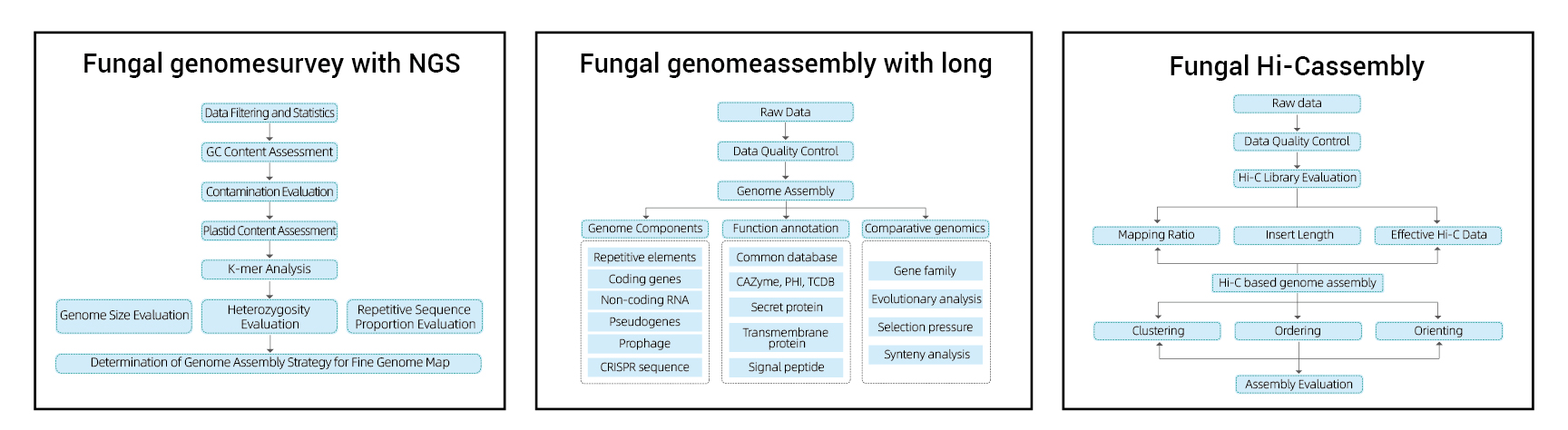
Genome Survey:
- Sequencing data quality control
- Genome estimation: size, heterozygosity, repetitive elements
Fine Genome Assembly:
- Sequencing Data Quality Control
- De novo Assembly
- Genome Component Analysis: Prediction of CDS and multiple genomic elements
- Functional annotation with multiple general databases (GO, KEGG, etc.) and advanced databases (CARD, VFDB, etc.)
Hi-C Assembly:
- Hi-C Library Assessment.
- Contigs anchoring clustering, ordering and orienting
- Evaluation of HI-C Assembly: Based on reference genome and heat map
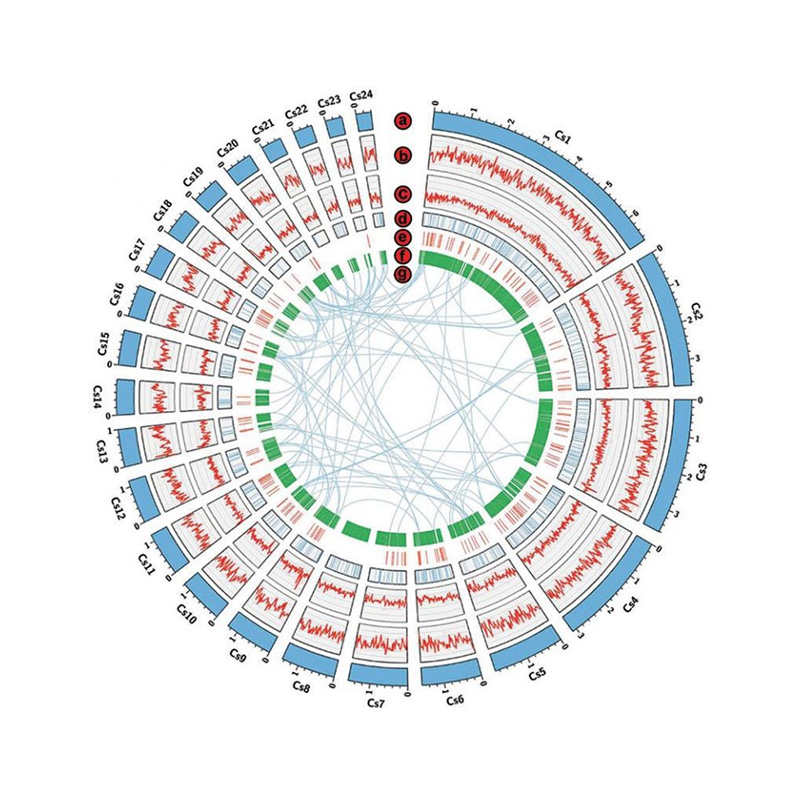
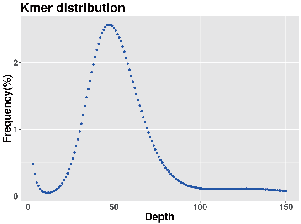
Genome survey: k-mer distribution
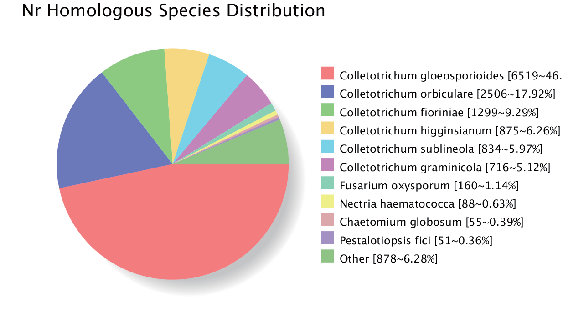
Genome assembly: gene homologous annotation (NR database)
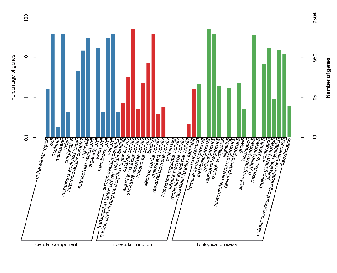
Genome assembly: functional gene annotation (GO)
Explore the advancements facilitated by BMKGene’s fungal genome assembly services through a curated collection of publications.
Hao, J. et al. (2023) ‘Integrated omic profiling of the medicinal mushroom Inonotus obliquus under submerged conditions’, BMC Genomics, 24(1), pp. 1–12. doi: 10.1186/S12864-023-09656-Z/FIGURES/3.
Lu, L. et al. (2023) ‘Genome sequencing reveals the evolution and pathogenic mechanisms of the wheat sharp eyespot pathogen Rhizoctonia cerealis’, The Crop Journal, 11(2), pp. 405–416. doi: 10.1016/J.CJ.2022.07.024.
Zhang, H. et al. (2023) ‘Genome Resources for Four Clarireedia Species Causing Dollar Spot on Diverse Turfgrasses’, Plant Disease, 107(3), pp. 929–934. doi: 10.1094/PDIS-08-22-1921-A
Zhang, S. S. et al. (2023) ‘Genetic and Molecular Evidence of a Tetrapolar Mating System in the Edible Mushroom Grifola frondosa’, Journal of Fungi, 9(10), p. 959. doi: 10.3390/JOF9100959/S1.