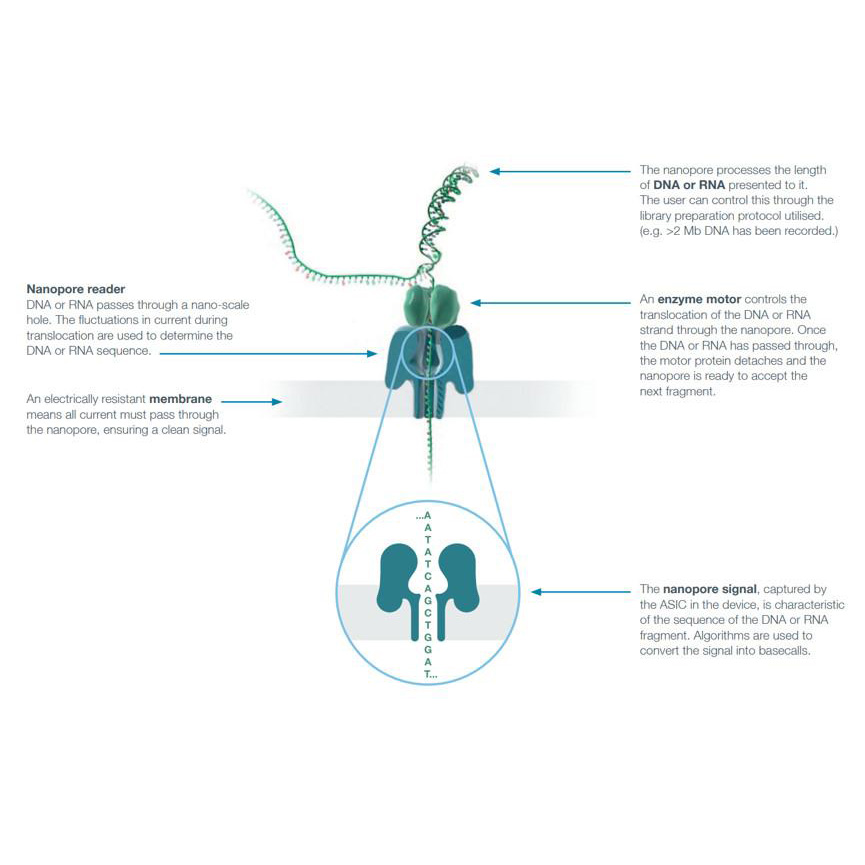
DNA/RNA Sequencing – Nanopore Sequencer
Service Details Features
|
Platform |
Library Size |
Theoretical Data Yield (Per Cell) |
Single-base Accuracy |
Applications |
|
Nanopore |
8Kb, 10kb, 20kb, Ultralong, cDNA-PCR |
70-90Gb/Cell |
85-92% |
SV calling, De novo, Full-length sequencing, Iso-Seq, Gene annotation, Detection of DNA methylation |
Service Advantages
● Over 5 years’ experience on PacBio sequencing platform with thousands of closed projects with various species.
● BMKGENE is an official partner of Oxford Nanopore, with dual platform RNA/DNA certification.
● There are mainstream models of sequencers with complete equipment and sufficient sequencing throughput.
● Based on the Nanopore platform, more than 10 animal and plant Denovo researches have been published in internationally renowned journals.
Sample Requirements
|
Sample Type |
Amount |
Concentrtion(Qubit ®) |
Volume |
Purity |
Others |
|
Genomic DNA |
Depend on Data Requirement |
≥20ng/μl |
≥15μl |
OD260/280=1.7-2.2; OD260/230≥1.5; Clear peak at 260 nm,no contaminations |
Concentration needs to be measured by Qubit and Qubit/Nanopore ≤ 2 |
|
Total RNA |
≥1.2μg |
≥100μg/μl |
≥15μl |
OD260/280=1.7-2.5; OD260/230=0.5-2.5;no contaminations |
RIN value ≥7.5 |
Service Workflow

Sample preparation

Library construction

Sequencing

Data analysis

Project delivery
Data Quality Assessment of DNA Sample
Table 1. Statistics on clean data.
|
BMKID |
rawSeqNum |
rawSumBase |
cleanSeqNum |
cleanSumBase |
cleanN50Len |
cleanN90Len |
cleanMeanLen |
cleanMaxLen |
cleanMeanQual |
|
DNA_BMK01 |
1,218,239 |
26.37 |
1,121,736 |
25.90 |
28,014 |
15,764 |
23,090 |
143,181 |
9 |
Data Quality Assessment of RNA Sample
Table 1. Statistics on clean data.
|
FileName |
ClientID |
ReadNum |
BaseNum |
N50 |
MeanLength |
MaxLength |
MeanQscore |
|
RNA_BMK001 |
C2 |
8,947,708 |
4,047,230,083 |
398 |
452 |
129,227 |
Q12 |
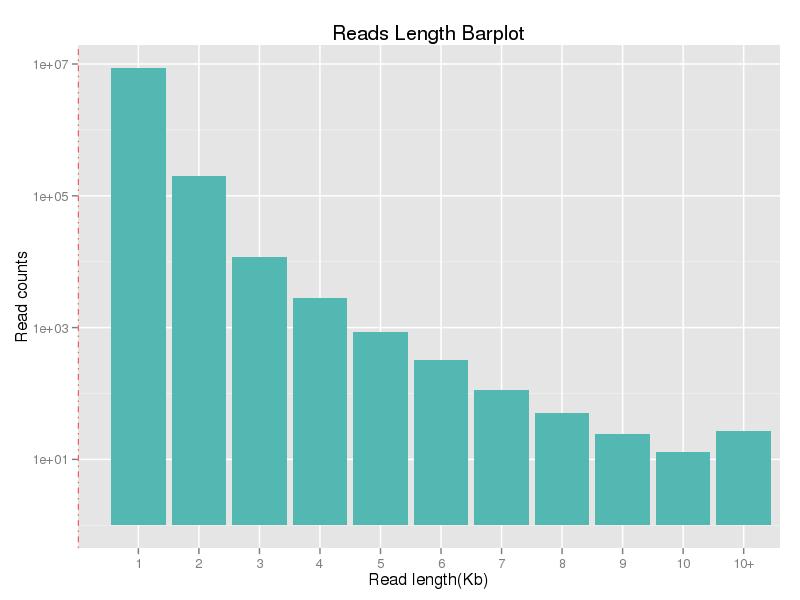
Figure 1. Read length distribution
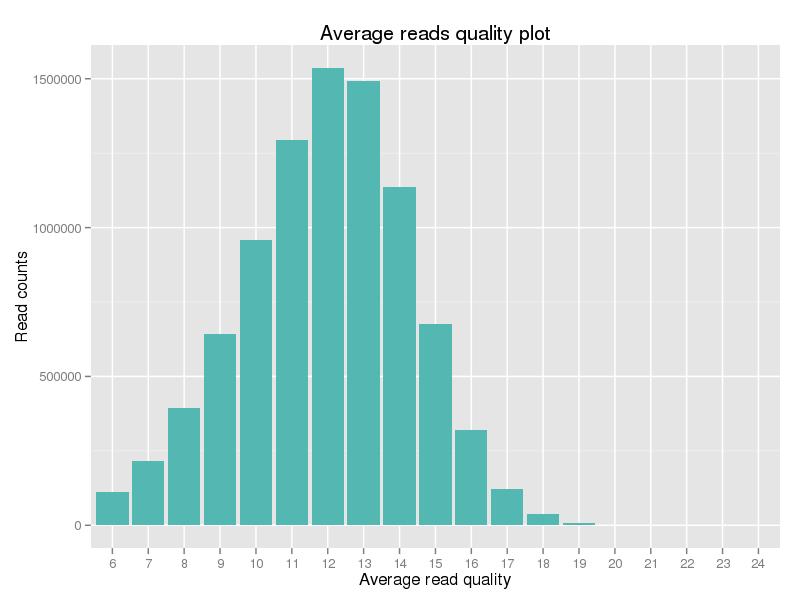
Figure 2. Quality score distribution of clean data
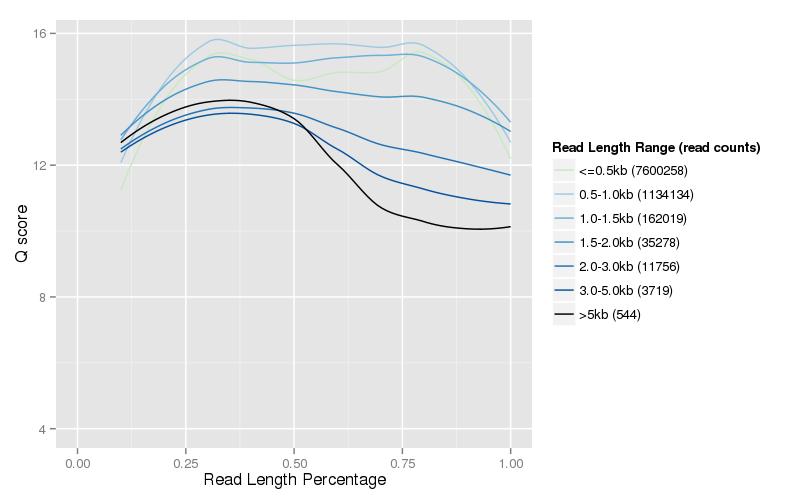
Figure 3. Length and quality score distribution of clean data