Bacterial and Fungal Whole Genome Re-Sequencing
Service Advantages
● Bioinformatic Analysis Includes Variant Calling: Providing functional insights into the re-sequenced genomes.
● Extensive Expertise: With thousands of microbial re-sequencing projects conducted annually, we bring over a decade of experience, a highly skilled analysis team, comprehensive content, and excellent post-sales support.
● Post-Sales Support: Our commitment extends beyond project completion with a 3-month after-sale service period. During this time, we offer project follow-up, troubleshooting assistance, and Q&A sessions to address any queries related to the results.
Service Specifications
|
Sequencing platform |
Sequencing Strategy |
Data recommended |
Quality control |
|
Illumina NovaSeq |
PE150 |
Depth of 100x |
Q30≥85% |
Sample Requirements
|
Concentration (ng/µL) |
Total amount (ng) |
Volume (µL) |
OD260/280 |
|
≥1 |
≥60 |
≥20 |
1.7-2.2 |
Bacteria: ≥1x107 cells
Unicellular Fungi: ≥5x106-1x107 cells
Macro Fungi: ≥4g
Service Work Flow

Sample delivery

Library construction

Sequencing

Data analysis

After-sale services
Includes the Following Analysis:
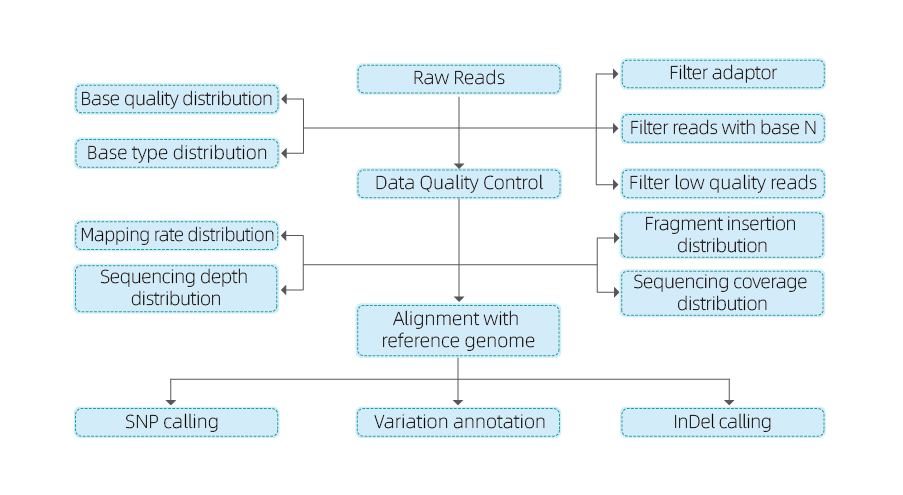
- Sequencing Data Quality Control
- Alignment to the Reference Genome
- Variant Calling: SNP and InDel
- Variant Annotation
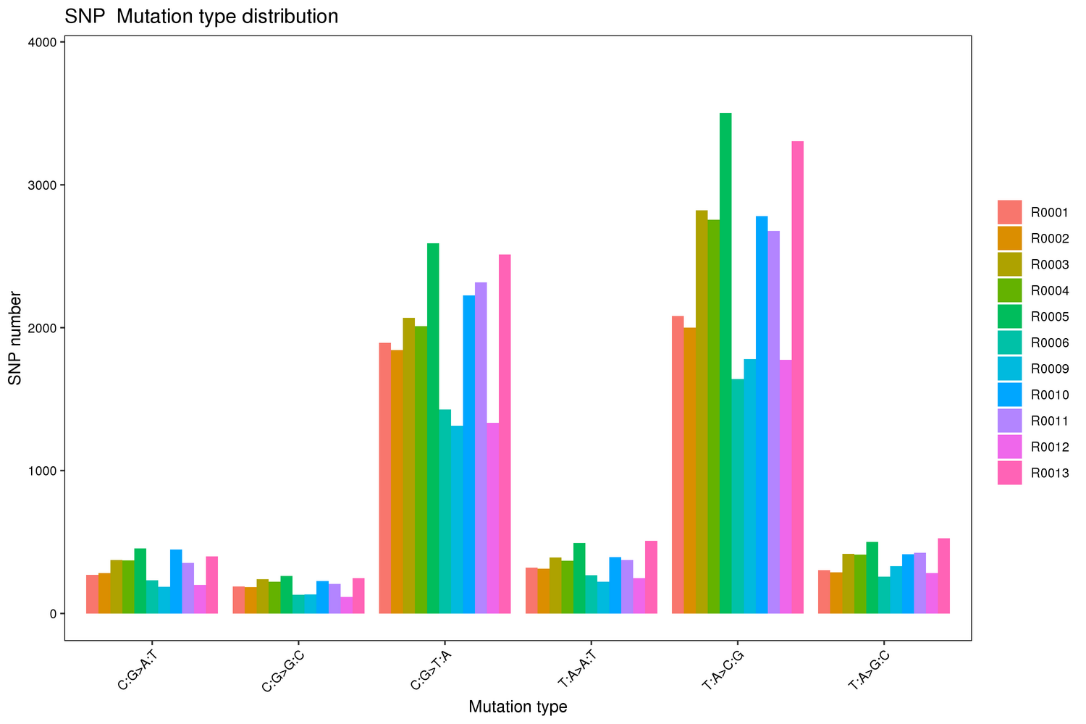
Variant calling: SNP types
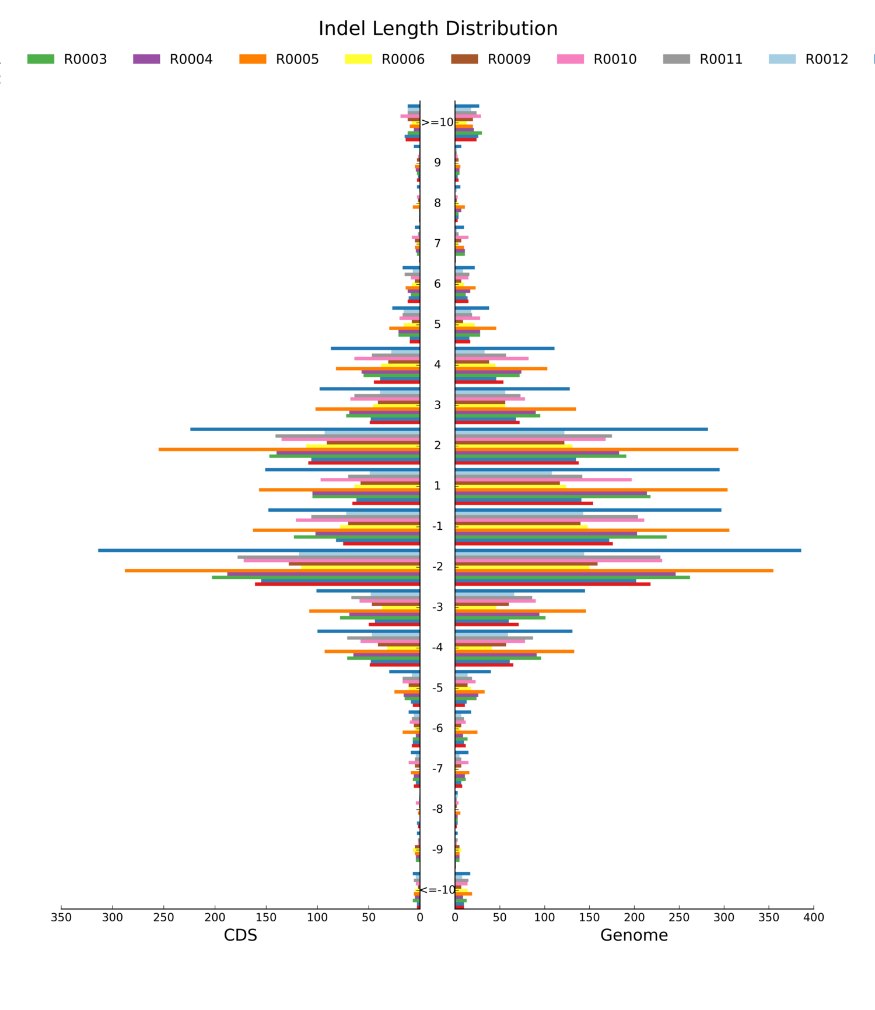
Variant calling: InDel length distribution
Explore the advancements facilitated by BMKGene’s microbial genome re-sequencing services through a curated collection of publications.
Jia, Y. et al. (2023) ‘Combining Transcriptome and Whole Genome Re-Sequencing to Screen Disease Resistance Genes for Wheat Dwarf Bunt’, International journal of molecular sciences, 24(24). doi: 10.3390/IJMS242417356.
Jiang, M. et al. (2023) ‘Ampicillin-controlled glucose metabolism manipulates the transition from tolerance to resistance in bacteria’, Science Advances, 9(10). doi: 10.1126/SCIADV.ADE8582/SUPPL_FILE/SCIADV.ADE8582_SM.PDF.
Yang, M. et al. (2022) ‘Aliidiomarina halalkaliphila sp. nov., a haloalkaliphilic bacterium isolated from a soda lake in Inner Mongolia Autonomous Region, China’, Int. J. Syst. Evol. Microbiol, 72, p. 5263. doi: 10.1099/ijsem.0.005263.
Zhu, Z., Wu, R. and Wang, G.-H. (2024) ‘Genome sequence of Staphylococcus nepalensis ZZ-2023a, isolated from Nasonia vitripennis’, Microbiology Resource Announcements. doi: 10.1128/MRA.00802-23.